Notice
Simulating the Extra Cellular Matrix - Calculations and data from atom to animal
- document 1 document 2 document 3
- niveau 1 niveau 2 niveau 3
Descriptif
Motivation:
The extracellular matrix (ECM) is a three-dimensional network of macromolecules that is the architectural support for cells and allows tissue cohesion. This dynamic structure regulates many biological functions such as adhesion, migration, proliferation, differentiation and cell survival.
From the modeling point of view, the ECM remains a challenging object to characterize : its study can be approached from different aspects and scales (quantum, atomic, molecular or even mesoscopic) which nevertheless remain complementary. Depending on the framework of investigation, the diversity of size, function and nature of the ECM molecules makes it necessary to choose a given scale of description. Indeed, there are still few methods available in physics that can describe/model a complex system with a unified approach combining physico-chemical properties linked to different scales of description.
Using simulations and modeling from the atomistic scale up to the mesoscopic scale as well as high-resolution imaging, we aim to understand and decipher the structural and dynamic behavior of ECM in healthy or pathological contexts.
Scientific and technical work:
The research projects developed within the MIME (modeling and multi-scale imaging) team (which is part of the MEDyC research unit) focus on a better description of the ECM, the interactions with its microenvironment as well as the impact of its modifications (natural such as the aging phenomenon, or pathological such as diabetic or cancer contexts). In particular, the following processes have been recently investigated:
(i) the detailed description of the interactions (through docking experiments and/or classical molecular dynamics (MD) simulations) between the various constituent elements of the ECM and molecules such as natural products.
(ii) the impact of post-translational modifications on ECM proteins structure, activity and dynamics through MD simulations at the atomic level.
(iii) the maturation of a modeling tool adapted to the mesoscopic scale of description.
Originality:
The strategy adopted by the MIME team relies on both theoretical and experimental biophysical approaches, whose results must be integrated in order to provide answers. This aggregation of knowledge takes the form of the DURABIN ((Developing Utilities for Nanometric Interactions in Biochemistry with Augmented Reality) mesoscopic integrator which, in the long run, should allow to see, understand and interact directly with the cell/microenvironment interface.
Results:
The localization of phenacetin at the plasma membrane was explored using extensive molecular dynamic simulations performed on several lipid bilayers. Insertion of phenacetin molecules into the membranes could be observed, and close examination of their position along the trajectory revealed that phenacetin adopted a specific orientation in the membrane with its amide group near lipid polar heads and its more deeply inserted ether group.
The effects of removal of sialic acid on the glycan, as well as on the dynamics of leucine-rich repeat L1 domain of the Insulin Receptor (IR) ectodomain were investigated through classical MD simulations. Perturbations in L1 domain dynamics were observed as a result of the removal of sialic acid. The perturbations include an increase in the flexibility of insulin-binding residues, which may affect insulin binding with IR. These changes are accompanied by perturbations in glycan–protein interactions and perturbation of long-range allosteric dynamics.
A set of tools has been developed within the DURABIN project (Developing Utilities for Nanometric Interactions in Biochemistry with Augmented Reality) to build models of large macromolecules or multimolecular complexes at the mesoscopic scale. Thus, models of individual full-length ECM proteins (i.e., collagen IV, laminin-111, and nidogen-1) and proteoglycan (perlecan) found in basement membranes, were built to generate a molecular network and assemble a three-dimensional model of a basement membrane-like ECM.
Impact:
The knowledge acquired from the investigations conducted at different scales improves the description of the different molecular actors and consolidates the evolution of the DURABIN simulation engine which is based on the concatenation of solid atomic and coarse-grained data sets. The use of this mesoscopic simulation tool should facilitate the interpretation of high-resolution imaging experiment and open the way to robust therapeutic strategies.
Choice of computing and storage resources used for this work:
MIME projects benefit from various computing facilities (ROMEO regional computing center, Jean Zay HPC computing center, CRIANN computing center) as well as the SOLEIL synchrotron facility. Atomic and coarse grained MD simulations are performed using GPU architecture. Generated and acquired data are stored on external disks and SSDS.
Bibliography:
Camille Fuseliet et al,
Low-diluted Phenacetinum disrupted the melanoma cancer cell migration
Scientific Reports, 9(1), 9109 (2019)
doi: 10.1038/s41598-019-45578-1.
Rajas M Rao et al,
Effects of changes in glycan composition on glycoprotein dynamics: Example of N-glycans on insulin receptor
Glycobiology, 31(9), 1121-1133 (2021)
doi: 10.1093/glycob/cwab049
Romain Rivet et al
Differential MMP-14 Targeting by Biglycan, Decorin, Fibromodulin and Lumican Unraveled by In Silico Approach.
AJP Cell Physiology, 324 (2), C353-C365 (2023)
doi: 10.1152/ajpcell.00429.2022
Hua Wong et al,
Multiscale modelling of the extracellular matrix.
Matrix Biology Plus, 13, 100096 (2022)
doi: 10.1016/j.mbplus.2021.100096
Thème
Documentation
Dans la même collection
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 5 : Des interactions croisées, à propos des compétences
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 2 : Les ateliers de la données
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 7 : Conclusion
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 4 : Des interactions croisées, liens entre le calcul et les données.
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 1 : Introduction et présentation des intervenants
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 6 : L'accompagnement autour de la gestion des données
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 3 : les liens entre les structures
-
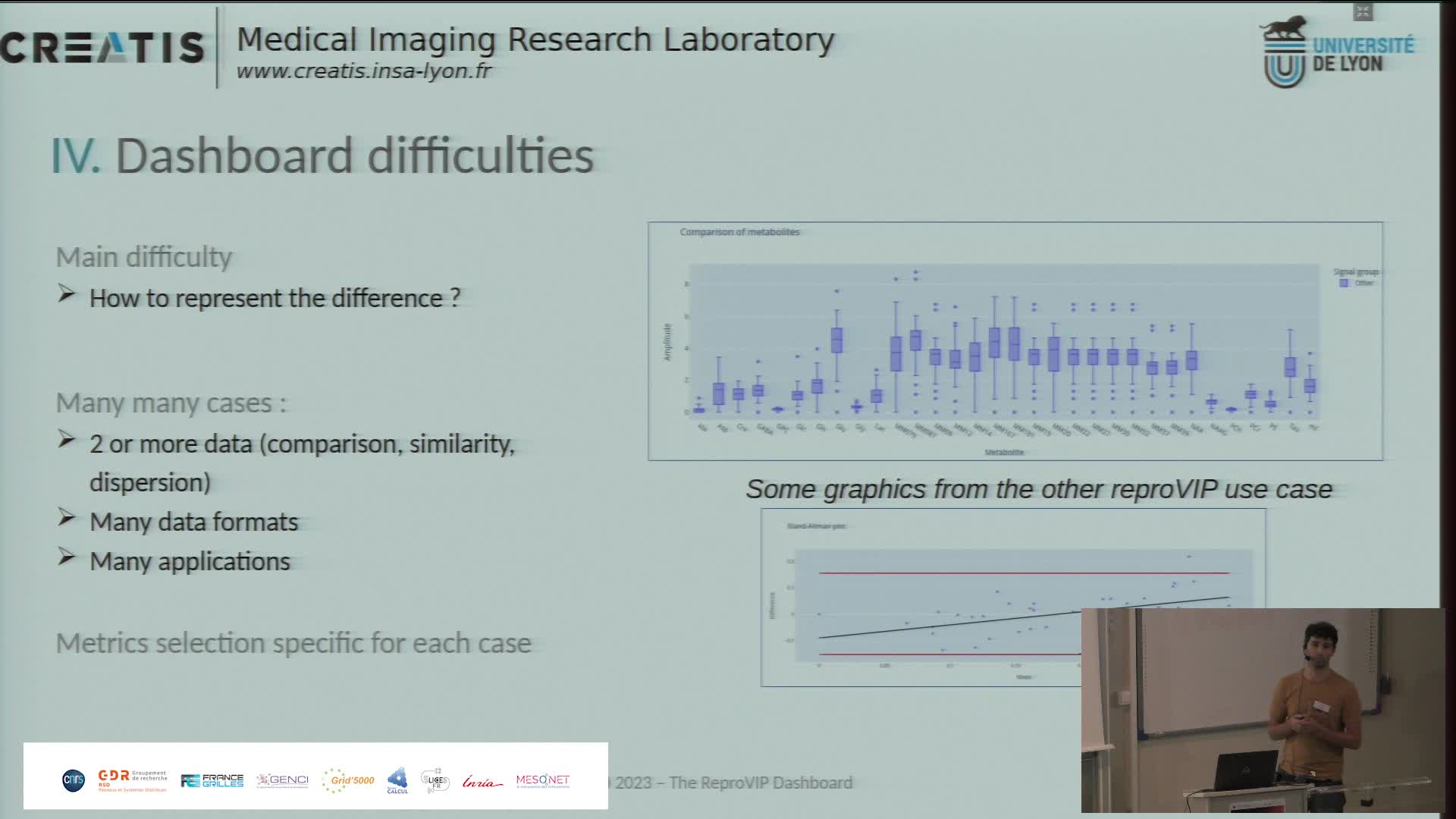
Présentation du dashboard ReproVIP pour visualiser la reproductibilité dans l'imagerie médicale
BonnetAxelLa plateforme d'imagerie virtuelle VIP [1] (https://vip.creatis.insa-lyon.fr) est un portail web de simulation et d'analyse d'images médicales. Elle existe depuis plus de 10 ans et a évolué pour
-
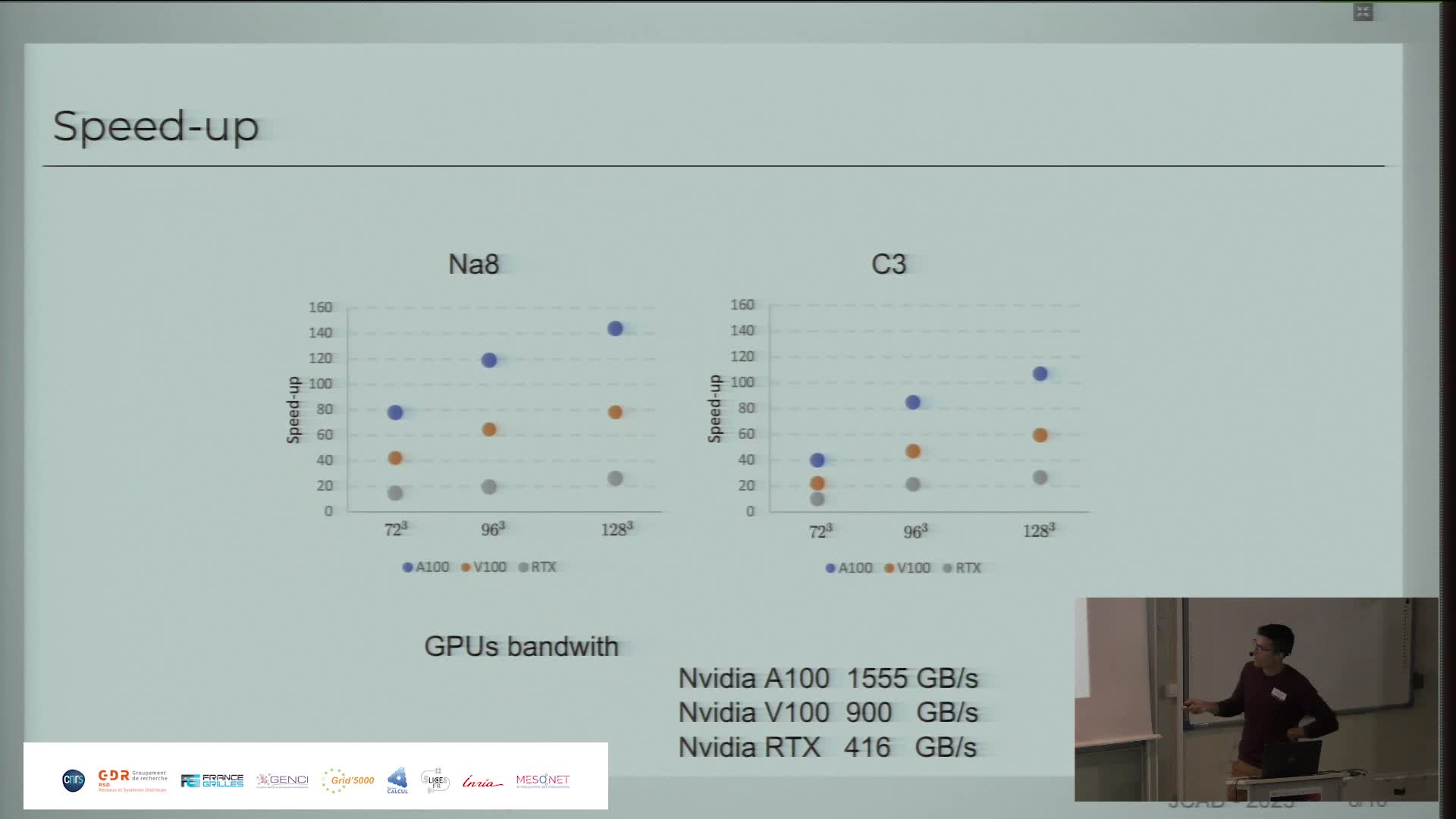
Présentation des performances paralléles du code QDD CUDA Fortran sur différentes Architectures GPU
PaipuriMahendraQDD est l'acronyme de Quantum Dissipative Dynamics, un ensemble de théories développées pour prendre en compte les corrélations dynamiques incohérentes dans les clusters et les molécules.
-
Parallélisation par l'intermédiaire d'une fenêtre à mémoire partagée (MPI 3.0) : application à un c…
ElyakimePierreJADIM est un code de calcul de mécanique des fluides développé en Fortran 90 à l'Institut de Mécanique des Fluides de Toulouse (IMFT).
-
Fast Polynomial Evaluation (présentation + demo)
VigneronFrançoisWe propose a new algorithm for quickly evaluating polynomials. The FPE algorithm pre-conditions a complex polynomial P of degree d in time O(d log d), with a low multiplicative constant independent
-
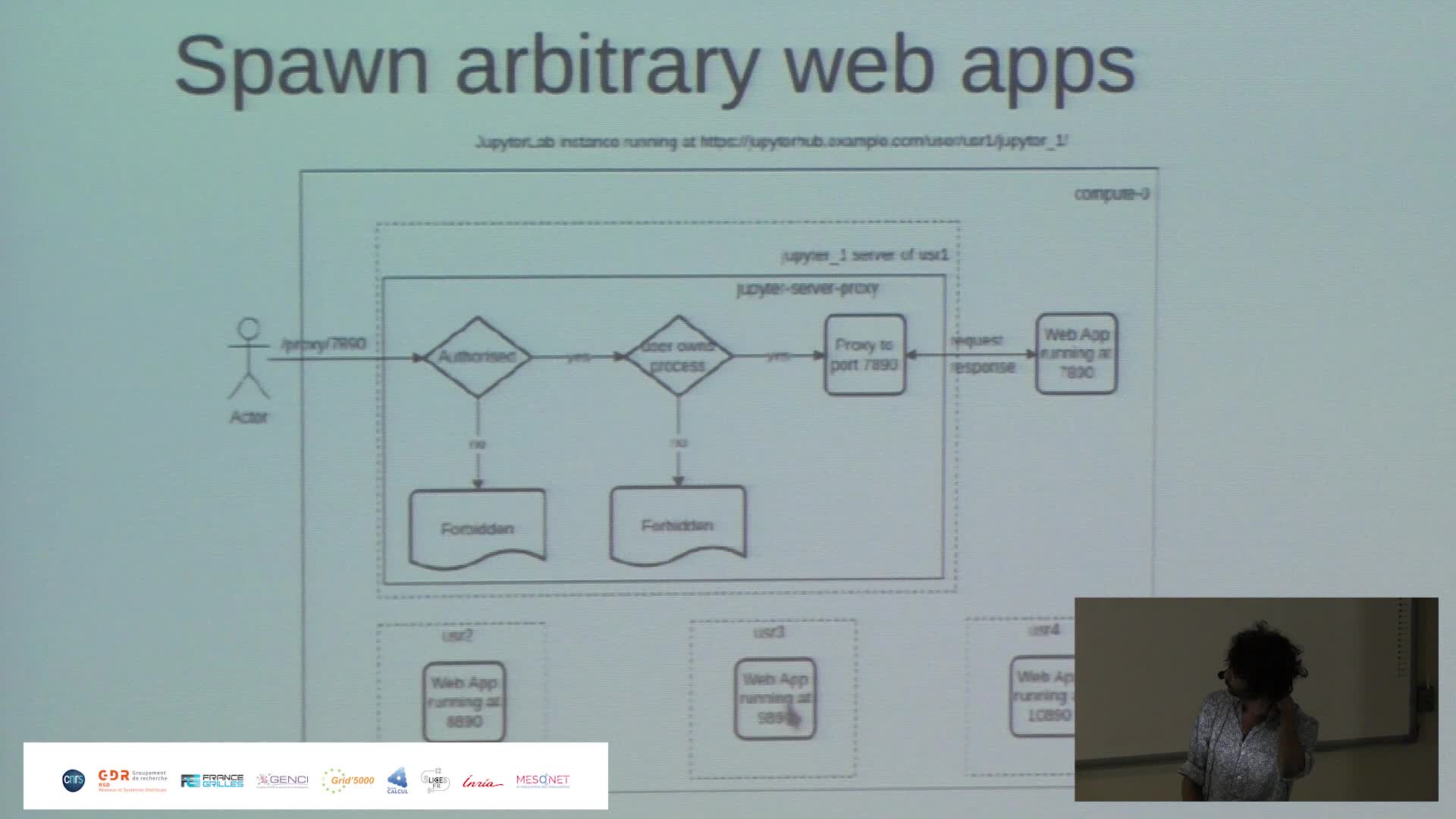
Harnessing the power of Jupyter{Hub,Lab} to make Jean Zay HPC resources more accessible
PaipuriMahendraThis talk revolves around the deployment of JupyterHub on Jean Zay HPC platform and how it is done to meet the demands of RSSI (ZRR constraints) and also to provide a seamless experience to the end