Notice
Using High Performance Computing to decipher the impact of ageing process on collagens
- document 1 document 2 document 3
- niveau 1 niveau 2 niveau 3
Descriptif
Motivations: According to the predictions made by The World Health Organization, between 2015 and 2050 the population over 60 years old will be doubled. But ageing is not related only to life expectancy. Biologically, ageing is defined as the accumulation of molecular and cellular damage.
One of the many reactions involved in ageing process is the carbamoylation. This reaction is the addition of isocyanic acid/cyanate to one specific amino acid, lysine, to form homocitrulline (Hct)(1). This modified residue can then be used as a biomarker of biological ageing (2). Collagens, the most abundant proteins in the human body accumulate Hct during ageing. High-performance computing (HPC) can be used to understand, at the molecular level, the structural and mechanical consequences of this important biomolecular process.
Scientific and technical work:
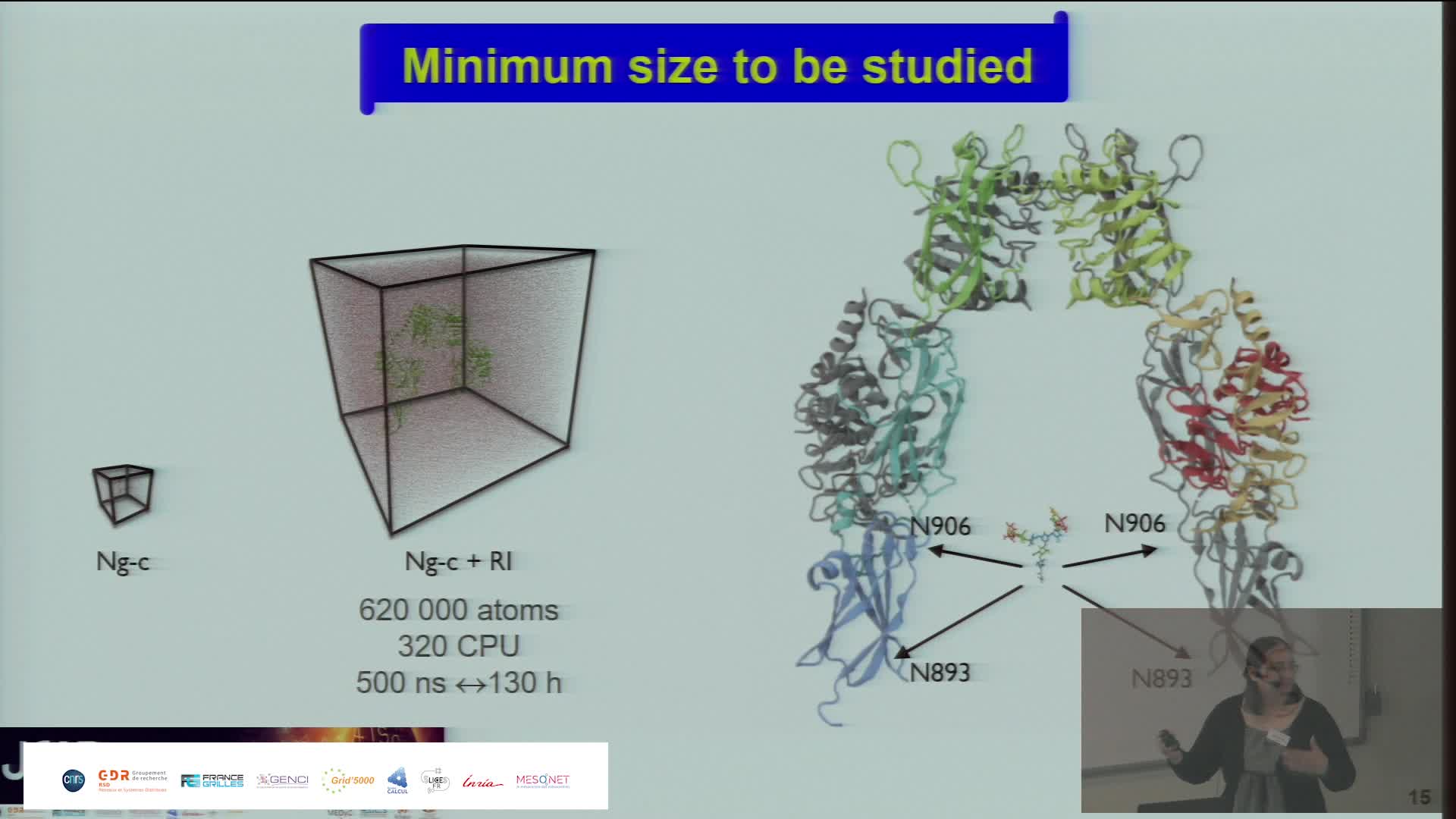
An in-silico model of collagen I was built. However, due to the huge size of the system (over 1000 amino acids per chain), MD simulations were performed only on smaller specific regions with Gromacs 2020.4 (3) and AMBER99SB*-ILDNP forcefield (4). Four different systems containing 0 to 3 Hct were investigated. Since there is no parameter to describe Hct in the AMBER forcefield, parametrization of this non-classical residue was done with quantum mechanics using HF/6-31G(d) level of theory. The trajectories obtained with the MD simulations were analysed with in-house scripts to evaluate the conformation of the Hct side chain (5,6).
Originality: The aim of the present PhD-thesis project is to decipher the molecular origins of the damages observed experimentally on collagen structures. Thus, the proposed strategy is a numerical approach mainly based on the use of molecular dynamics (MD) as a powerful computational approach applied to collagen I. MD calculations act as microscopic probe that can provide insights about the structure and dynamics of Hct. This atomic characterization can explain the behaviours encountered at macroscopic level on collagen I.
Results: The data obtained from computationally demanding MD simulations allowed us to characterize the Hct side chain behaviour and to compare it to the behaviour of the lysine side chain. On collagen, differences were detected between the dihedral angles' population of lysine and the one of Hct residue and could explain the loss of certain collagen I properties observed experimentally in our research unit.
Impact: Thanks to this work, the disruption of key interactions involving the site of carbamoylation could be identified. By extension, a link could be made with some behaviour observed at the macroscopic level.
Choice of computing and storage resources used for this work:
For this work, 1 node was used with the following specifications: 4 MPI tasks, 7 OpenMP threads per MPI task and 4 GPUs per node. Due to the size of the system, using more computing resources was not relevant. But in the future, since the size of the system will be increased, more nodes will be required for the computations. All data were stored on external disks and SSDS.
Bibliography:
1. Jaisson S, Pietrement C, Gillery P. Carbamylation-Derived Products: Bioactive Compounds and Potential Biomarkers in Chronic Renal Failure and Atherosclerosis. Clinical Chemistry. 1 nov 2011;57(11):1499‑505.
2. Gorisse L, Pietrement C, Vuiblet V, Schmelzer CEH, Köhler M, Duca L, et al. Protein carbamylation is a hallmark of aging. Proc Natl Acad Sci USA. 2 févr 2016;113(5):1191‑6.
3. Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. J Comput Chem. déc 2005;26(16):1701‑18.
4. Aliev AE, Kulke M, Khaneja HS, Chudasama V, Sheppard TD, Lanigan RM. Motional timescale predictions by molecular dynamics simulations: Case study using proline and hydroxyproline sidechain dynamics: Proline Force Field Parameters. Proteins. févr 2014;82(2):195‑215.
5. Dunbrack RL, Karplus M. Backbone-dependent Rotamer Library for Proteins Application to Side-chain Prediction. Journal of Molecular Biology. 1993;230(2):543‑74.
6. Shapovalov MV, Dunbrack RL. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure. juin 2011;19(6):844‑58.
Intervention / Responsable scientifique
Thème
Documentation
Dans la même collection
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 1 : Introduction et présentation des intervenants
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 6 : L'accompagnement autour de la gestion des données
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 3 : les liens entre les structures
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 5 : Des interactions croisées, à propos des compétences
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 2 : Les ateliers de la données
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 7 : Conclusion
-
Table ronde et discussions : infrastructures de calcul et ateliers de la donnée de recherche Data G…
CastexStéphanieRenardArnaudAlbaretLucieRenonNicolasDufayardJean-FrançoisPARTIE 4 : Des interactions croisées, liens entre le calcul et les données.
-
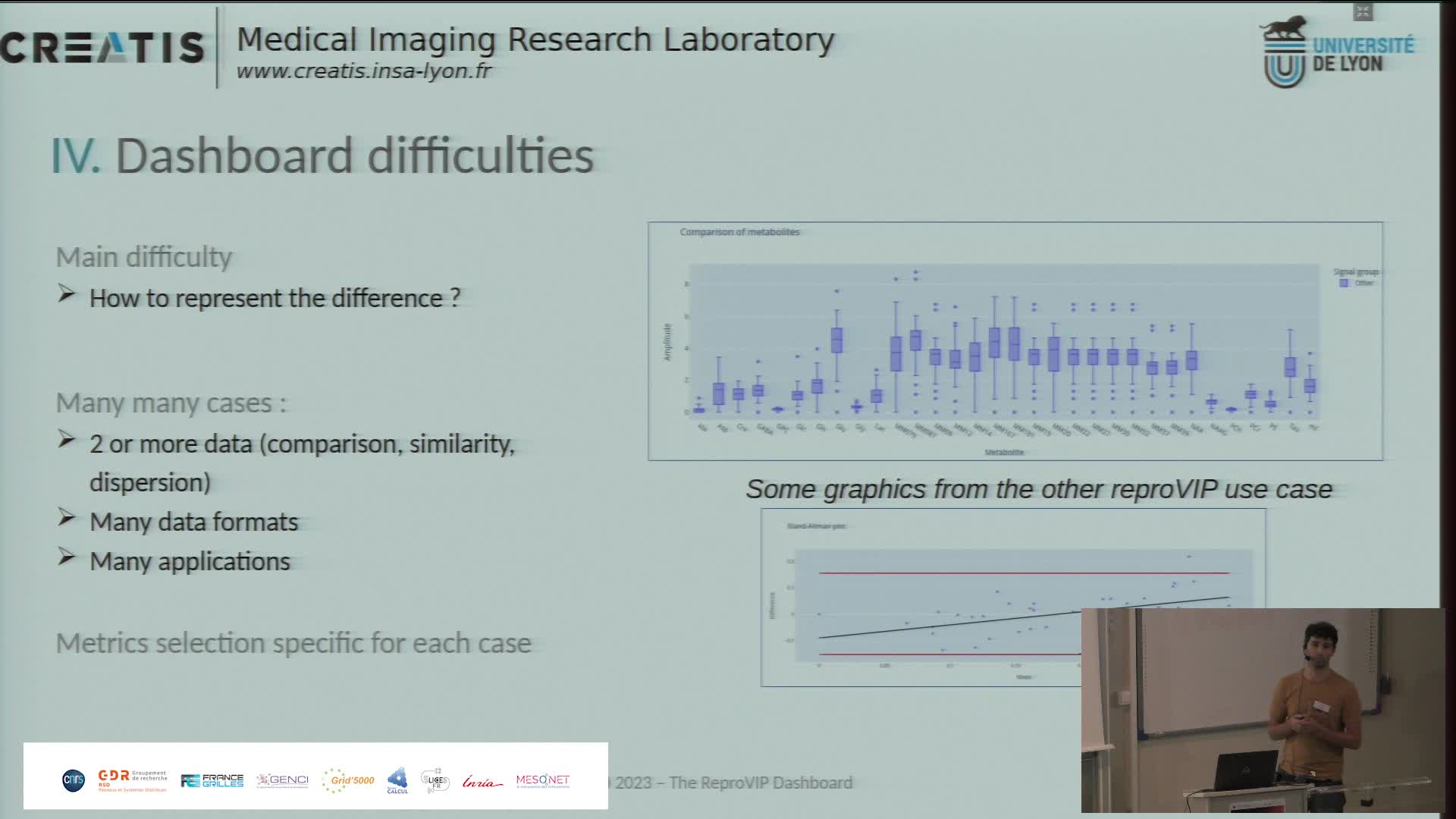
Présentation du dashboard ReproVIP pour visualiser la reproductibilité dans l'imagerie médicale
BonnetAxelLa plateforme d'imagerie virtuelle VIP [1] (https://vip.creatis.insa-lyon.fr) est un portail web de simulation et d'analyse d'images médicales. Elle existe depuis plus de 10 ans et a évolué pour
-
Parallélisation par l'intermédiaire d'une fenêtre à mémoire partagée (MPI 3.0) : application à un c…
ElyakimePierreJADIM est un code de calcul de mécanique des fluides développé en Fortran 90 à l'Institut de Mécanique des Fluides de Toulouse (IMFT).
-
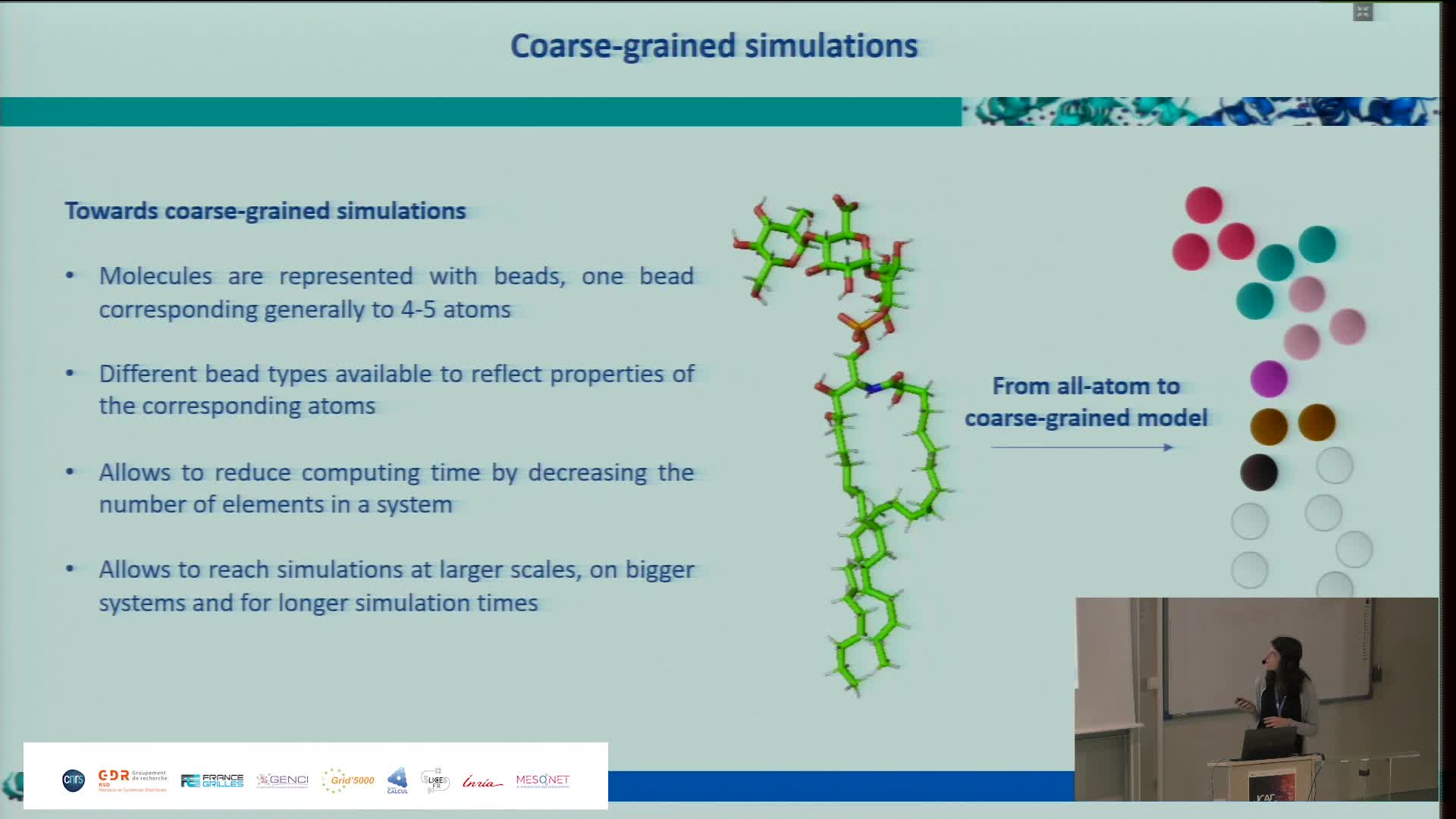
Simulating the Extra Cellular Matrix - Calculations and data from atom to animal
BaudStéphanieThe extracellular matrix (ECM) is a three-dimensional network of macromolecules that is the architectural support for cells and allows tissue cohesion. This dynamic structure regulates many biological
-
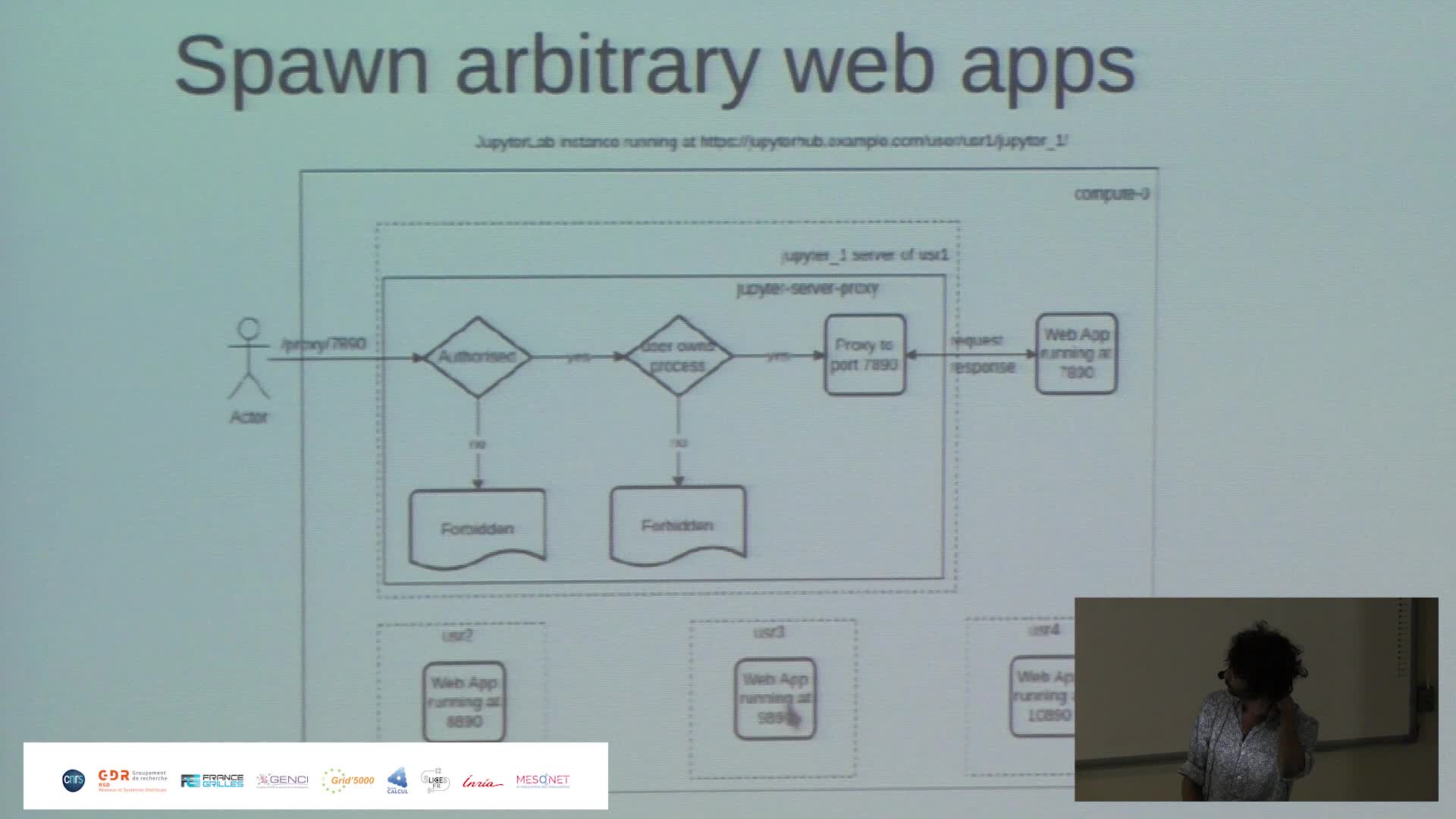
Harnessing the power of Jupyter{Hub,Lab} to make Jean Zay HPC resources more accessible
PaipuriMahendraThis talk revolves around the deployment of JupyterHub on Jean Zay HPC platform and how it is done to meet the demands of RSSI (ZRR constraints) and also to provide a seamless experience to the end
-
Using High Performance Computing to decipher the impact of ageing process on collagens
DepenveillerCamillePlant cells have to face environmental stress, and in this context, being able to decipher the organization of the plant plasma membrane is a key point to better understand this adaptability of plasma