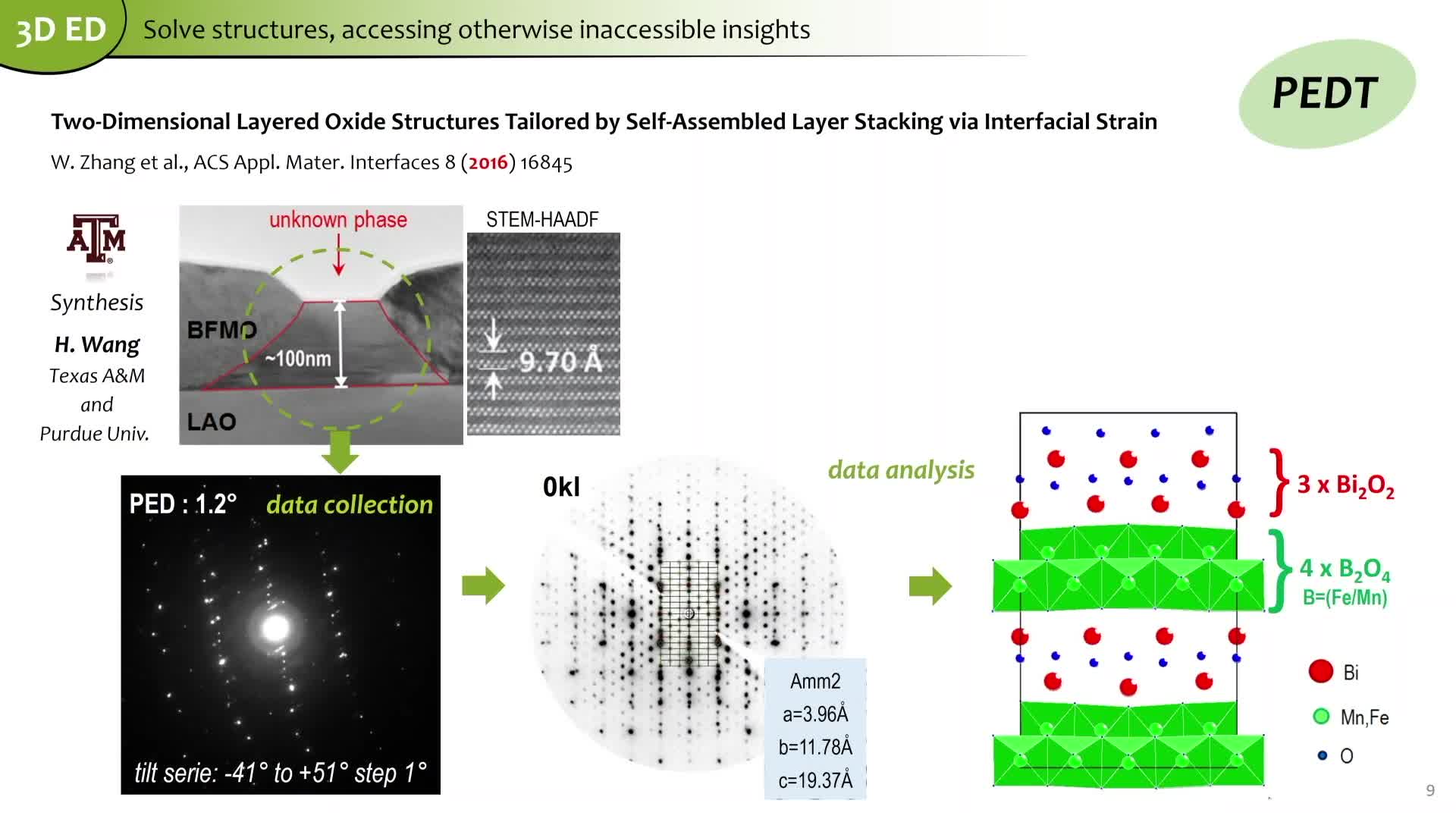
Notice
Fast adaptive optics fluorescence microscopy for in vivo high resolution imaging in depth
- document 1 document 2 document 3
- niveau 1 niveau 2 niveau 3
Descriptif
The study of fast biological phenomena at the cellular level has become possible even in depth in biological tissues in several animal models and rodents, thanks to efficient 3D microscopy techniques. Light sheet fluorescence microscopy (LSFM) is particularly well suited to imaging relatively transparent specimens such as zebrafish, while 2-photon excited fluorescence microscopy (TPEF) remains the technique of choice for more scattering tissues like mammalian brain. But image quality deep inside biological tissues remains limited by scattering and optical aberrations due to the refractive index inhomogeneity, which limit both resolution and sensitivity in all microscopy modalities.
A novel approach for fast adaptive optics (AO) in fluorescence microscopy. To overcome this difficulty, in particular regarding aberrations, AO has been implemented on several linear and non-linear optical sectioning microscopy setups and currently provides a reliable live correction of the aberrations, enabling functional imaging of synaptic boutons, axons and spines and soma in infragranular layers of the mouse cortex in 2-photon microscopy, or enabling subcellular imaging of organelle dynamics in the early zebrafish brain in light-sheet microscopy. Significant gain in spatial resolution and signal intensity was demonstrated, but the two approaches based on a direct wavefront sensing or a sensorless process still suffer from several limitations, linked to the complex generation or the invasive introduction of a guide star inside the sample, or a time-consuming iterative approach to reach a good correction. We proposed a few years ago an innovative strategy of adaptive optics for neuroimaging, by adapting pioneer work from astronomy to the constraints of fluorescence microscopy.
By using an extended-source Shack-Hartmann wavefront sensor (ESSH), our approach allows to preserve speed and accuracy of the direct wavefront sensing method and takes advantage of existing labelling methods of biological samples [1]. It relies on the cross-correlation of images of an extended source obtained through a microlens array and requires to be coupled to an optical sectioning method in order to provide a guide plane: it is thus interestingly compatible with various fluorescence imaging techniques already implemented for 3D imaging such as two-photon microscopy [2] or light-sheet fluorescence microscopy [3].
AO-enhanced TPEF and LSFM for biological imaging in depth. We have implemented this original wavefront sensor into a home-built adaptive optics two-photon setup (AOTPEF) with a two-color labeling strategy: the wavefront is measured thanks to a red anatomical label, thus preserving the photon budget of the green label of interest. We have shown the benefit of the extended scene Shack-Hartmann wavefront measurement in the case of scattering samples, and its improved accuracy at large depths under low signal-to-background ratio conditions, enabling deeper direct wavefront measurement and AO in biological tissues (typically up to 4 mean free paths) [2]. As shown on the figure below, we have demonstrated the ability to correct aberrations up to 350 microns deep inside a fixed cortical brain slice from GAD-GFP mice, with labeled inhibitory neurons, proving large gain in intensity and resolution, with an increased contrast by a typical factor of 5 for small structures. Since the scattering length in fixed brain tissue is two times smaller than in intact tissue, the technique promises to provide a strong advantage for in vivo imaging down to significant depths (≥ 700µm in the mouse brain). We also performed ESSH-based AO integration into LSFM microscopy, in order to quantitatively demonstrate the performance of the new, fast closed-loop approach. We developed two homebuilt AO-LSFM setups: 1) an AO-DSLM setup (DSLM for Digitally Scanned Light-sheet fluorescence microscopy), including AO in the emission path, in order to correct for sample-induced aberrations, and 2) an AOASLM setup (ASLM for Axially-Swept Light-Sheet Microscopy), also with AO in the emission path, for highresolution 3D imaging in densely labeled samples. Based on two-color labeling of the sample, which is routinely used in biology to enable structural and functional imaging, we then demonstrated ESSH-based AO running at 10Hz with an optimal photon budget, allowing improved imaging of GCaMP7b labeled neurons in depth in the live adult Drosophila brain using AO-DSLM [3]. Recently, we have shown that ESSH-based AO-ASLM provides diffraction-limited 3D resolution deep (>300µm) into the zebrafish brain, by correcting sample-induced aberrations, together with an x2 signal increase for large cells, and maintaining the ability to distinguish individual cells even at great depths. This is key to many studies, as a major biomarker is the quantification of cell amount/density (segmentation) in many fields (developmental biology, neuro, cardio).
References
[1] Hubert, A.et al. Adaptive optics light-sheet microscopy based on direct wavefront sensing without any guide star, Opt. Lett44(10) 2514-2517 (2019)
[2] Imperato, S. et al. Single-shot quantitative aberration and scattering length measurements in mouse brain tissues using an extended-source Shack-Hartmann wavefront sensor, Optics Express 30(9)15250-15265 (2022)
[3] Hubert, A. et al. Enhanced neuroimaging with a calcium sensor in the live adult Drosophila Melanogaster brain using closed-loop adaptive optics light-sheet microscopy, Journal of Biomedical Optics, 28, 6 (2023)
Sur le même thème
-
Super-resolved fluorescence microscopy using random illuminations (RIM)
SentenacAnneLe GoffLoïcThe resolution in optical microscopy is fundamentally constrained by the diffraction limit…
-
Imagerie structurelle 3D de tissus riches en collagène par microscopie SHG polarimétrique
Schanne-KleinMarie-ClaireLa microscopie par génération de seconde harmonique (SHG) permet d’imager le collagène.…
-
Impact des techniques neuronales pour la microscopie computationnelle
DorizziBernadetteGottesmanYaneckCet exposé s'attachera à décrire l'intérêt des techniques de réseaux de neurones pour la microscopie computationnelle…
-
Des traces de vie sur l'astéroïde Bénou ? | OSIRIS-REx
ZanettaPierre-MarieLes premiers résultats du retour d'échantillons de la mission OSIRIS-REx suite à son exploration de l'astéroïde Bénou.
-
La vie microscopique dans les arts vivants des 20e et 21e siècles (partie 1)
SégingerGisèleHeyraudViolaineShepherd-BarrKirstenAbdallaDaniel IbrahimPrateekArmstrongPatrickDarrobersElisabethBazinWallerandCette rencontre est organisée par Liliane Campos, François-Joseph Lapointe et Kirsten Shepherd-Barr, dans le cadre du projet BioCriticism avec le soutien du projet Biohumanities (dir. Gisèle Séginger)
-
-
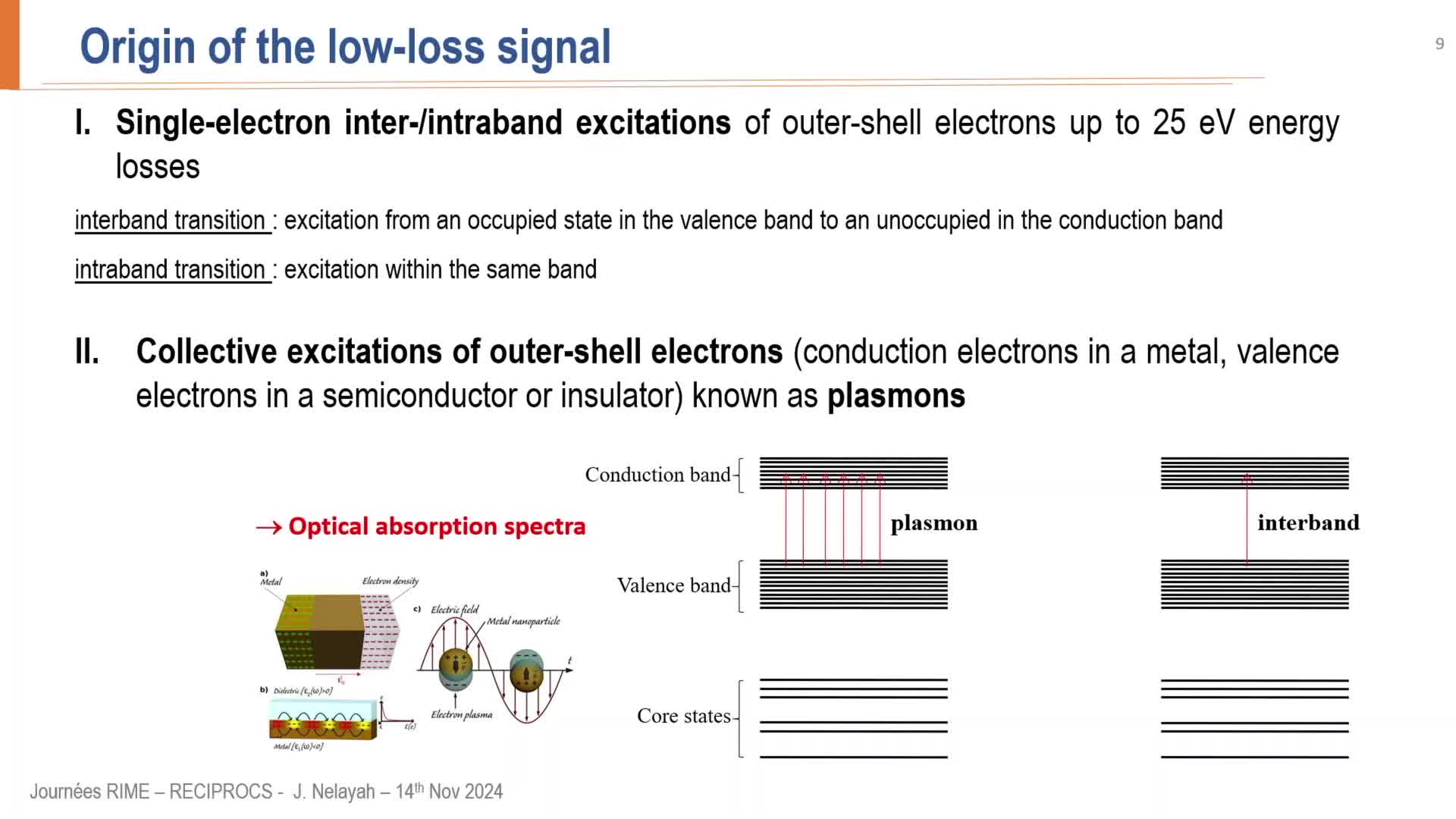
EELS, principe et application à l’étude des nanomatériaux inorganiques
NelayahJaysenEELS, principe et application à l’étude des nanomatériaux inorganiques
-
-
Nanoscale structural heterogeneity of crystalline carbohydrates
OgawaYuNanoscale structural heterogeneity of crystalline carbohydrates
-
-
Actualités astronomiques de juillet 2023
PécontalEmmanuelExtremely Large Telescope ; suite de la mission DART ; exoplanètes troyennes ; Rho Ophiuchi et Saturne vues par le JWST et grains carbonés dans une galaxie lointaine : les actualités astronomiques de
-
Les étoiles et leur environnement proche observés à haute résolution angulaire
KervellaPierreDepuis les très faibles naines brunes jusqu'aux immenses supergéantes, les étoiles sont le moteur de l'évolution chimique de l'Univers. Souvent variables, binaires ou multiples, elles présentent une