Rechenmann, François (19..-.... ; informaticien)
Ingénieur et Docteur-Ingénieur en informatique, François Rechenmann est chercheur au centre Inria Grenoble – Rhône-Alpes. Il y exerce ses activités à l’interface de l’informatique et des sciences du vivant en contribuant plus particulièrement au développement de méthodes et de logiciels pour l’analyse des séquences génomiques des microorganismes. Cofondateur de la société Genostar, qui propose des solutions bioinformatiques aux industries pharmaceutiques, agroalimentaires et biotechnologiques, il en est le conseiller scientifique. Très impliqué dans les actions de médiation scientifique, François Rechenmann est le responsable scientifique du site Interstices dont l’objectif est d’expliquer l’informatique en tant que domaine de recherche.
Vidéos
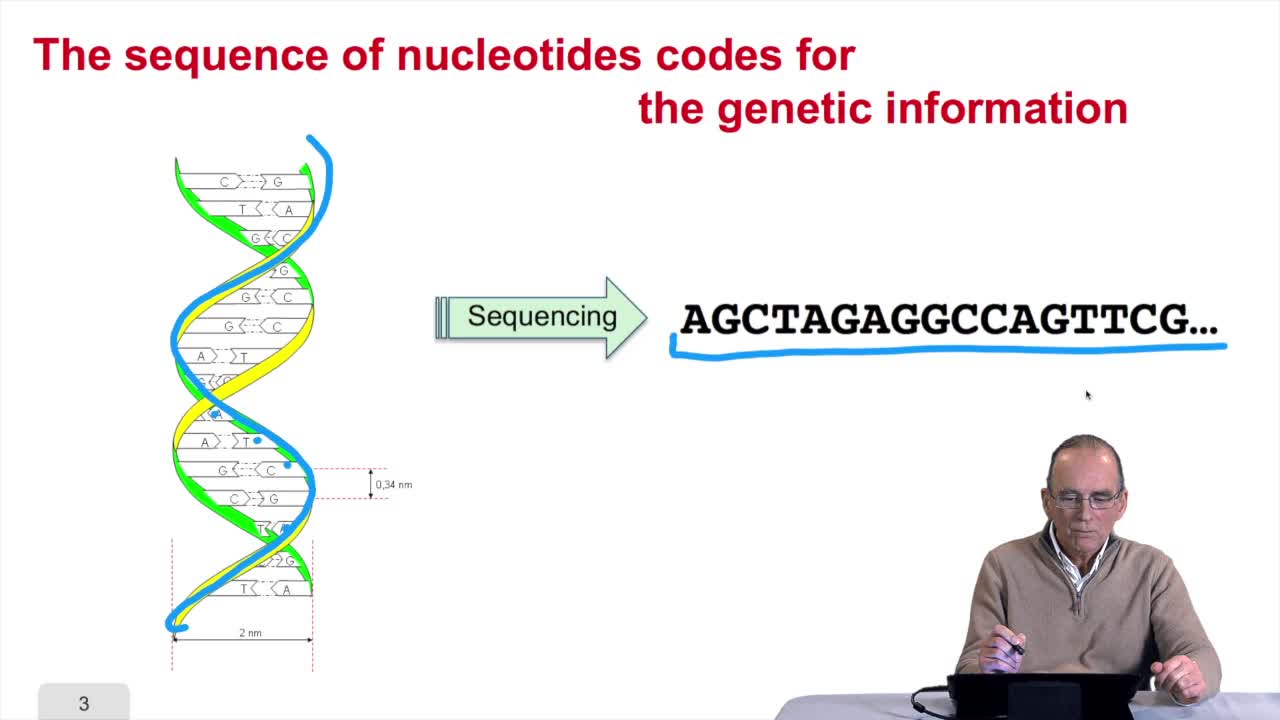
1.3. DNA codes for genetic information
Remember at the heart of any cell,there is this very long molecule which is called a macromolecule for this reason, which is the DNA molecule. Now we will see that DNA molecules support what is called
2.1. The sequence as a model of DNA
Welcome back to our course on genomes and algorithms that is a computer analysis ofgenetic information. Last week we introduced the very basic concept in biology that is cell, DNA, genome, genes
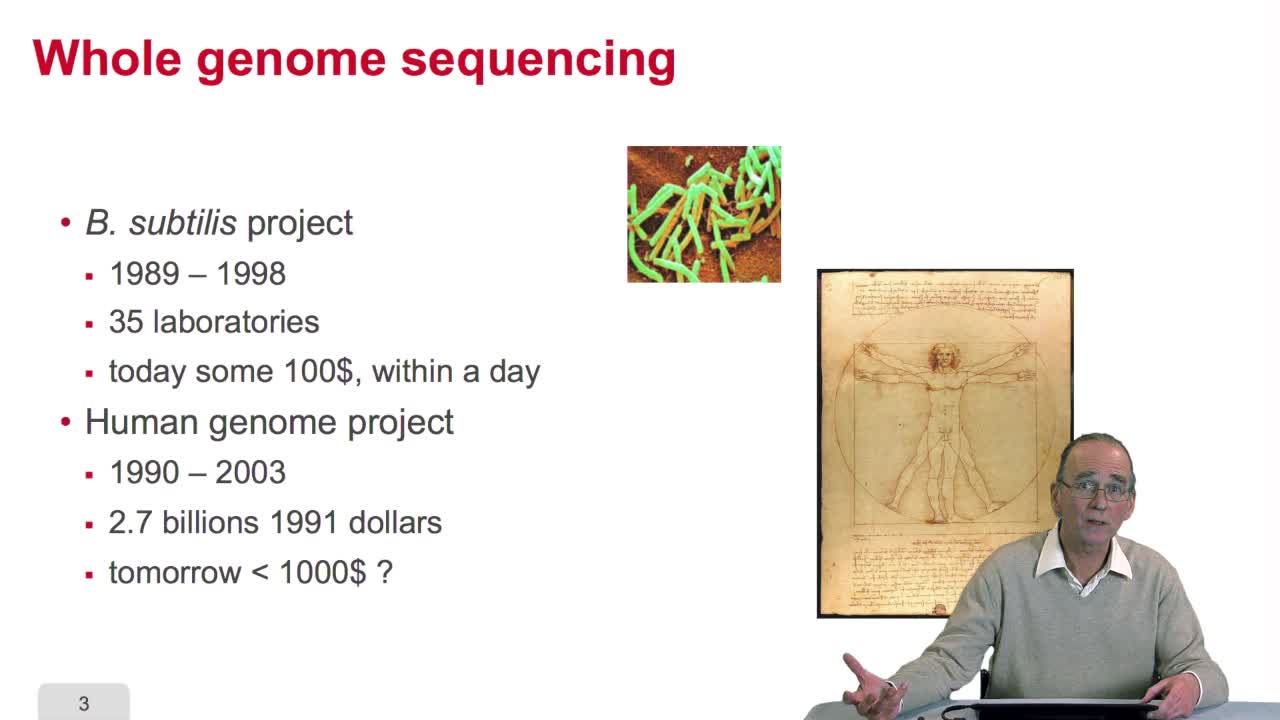
2.9. Whole genome sequencing
Sequencing is anexponential technology. The progresses in this technologyallow now to a sequence whole genome, complete genome. What does it mean? Well let'stake two examples: some twenty years ago,
3.7. Index and suffix trees
We have seen with the Boyer-Moore algorithm how we can increase the efficiency of spin searching through the pre-processing of the pattern to be searched. Now we will see that an alternative way of
4.4. Aligning sequences is an optimization problem
We have seen a nice and a quitesimple solution for measuring the similarity between two sequences. It relied on the so-called hammingdistance that is counting the number of differencesbetween two
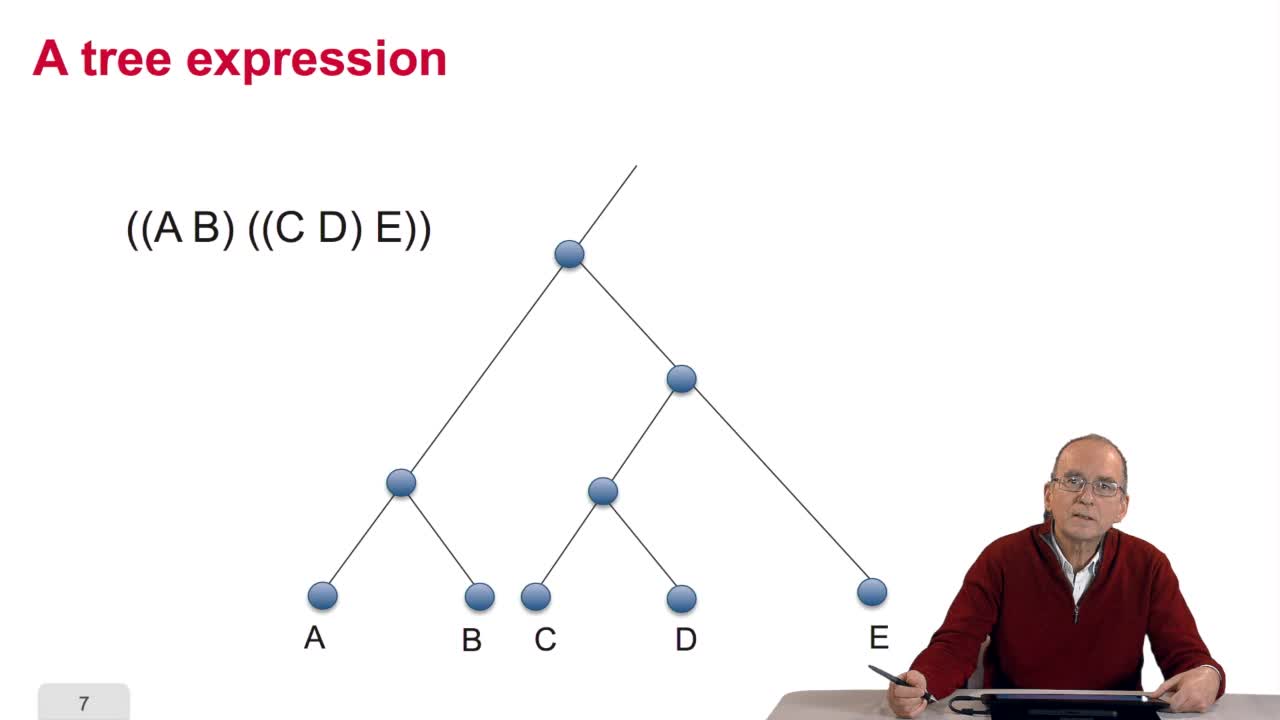
5.2. The tree, an abstract object
When we speak of trees, of species,of phylogenetic trees, of course, it's a metaphoric view of a real tree. Our trees are abstract objects. Here is a tree and the different components of this tree.
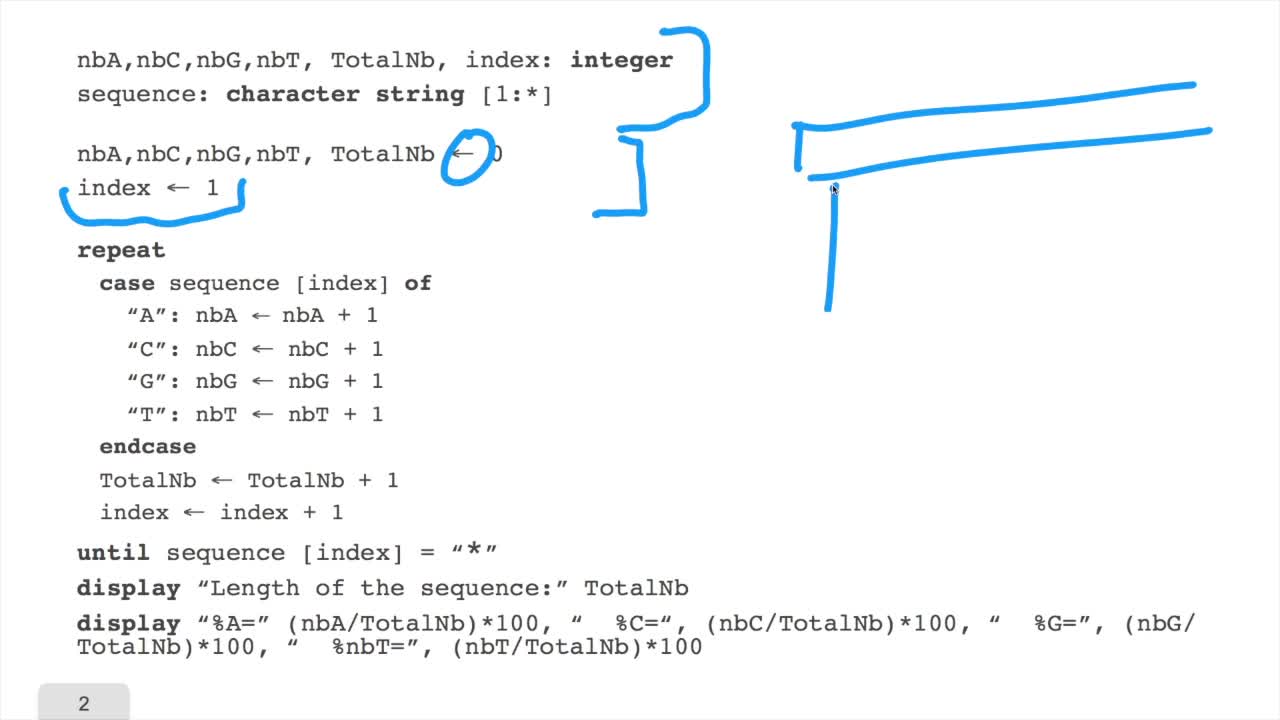
1.6. GC and AT contents of DNA sequence
We have designed our first algorithmfor counting nucleotides. Remember, what we have writtenin pseudo code is first declaration of variables. We have several integer variables that are variables which
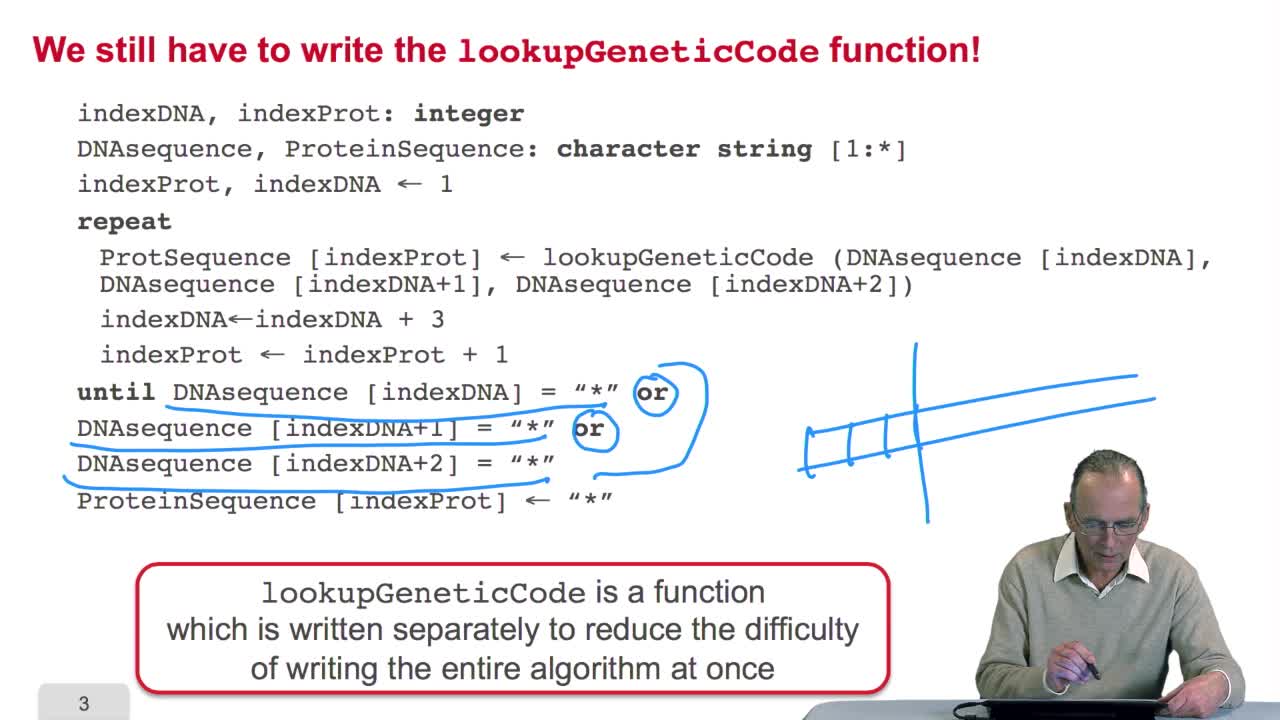
2.5. Implementing the genetic code
Remember we were designing our translation algorithm and since we are a bit lazy, we decided to make the hypothesis that there was the adequate function forimplementing the genetic code. It's now time
3.2. A simple algorithm for gene prediction
Based on the principle we statedin the last session, we will now write in pseudo code a firstalgorithm for locating genes on a bacterial genome. Remember first how this algorithm should work, we first
3.10. Gene prediction in eukaryotic genomes
If it is possible to have verygood predictions for bacterial genes, it's certainly not the caseyet for eukaryotic genomes. Eukaryotic cells have manydifferences in comparison to prokaryotic cells. You
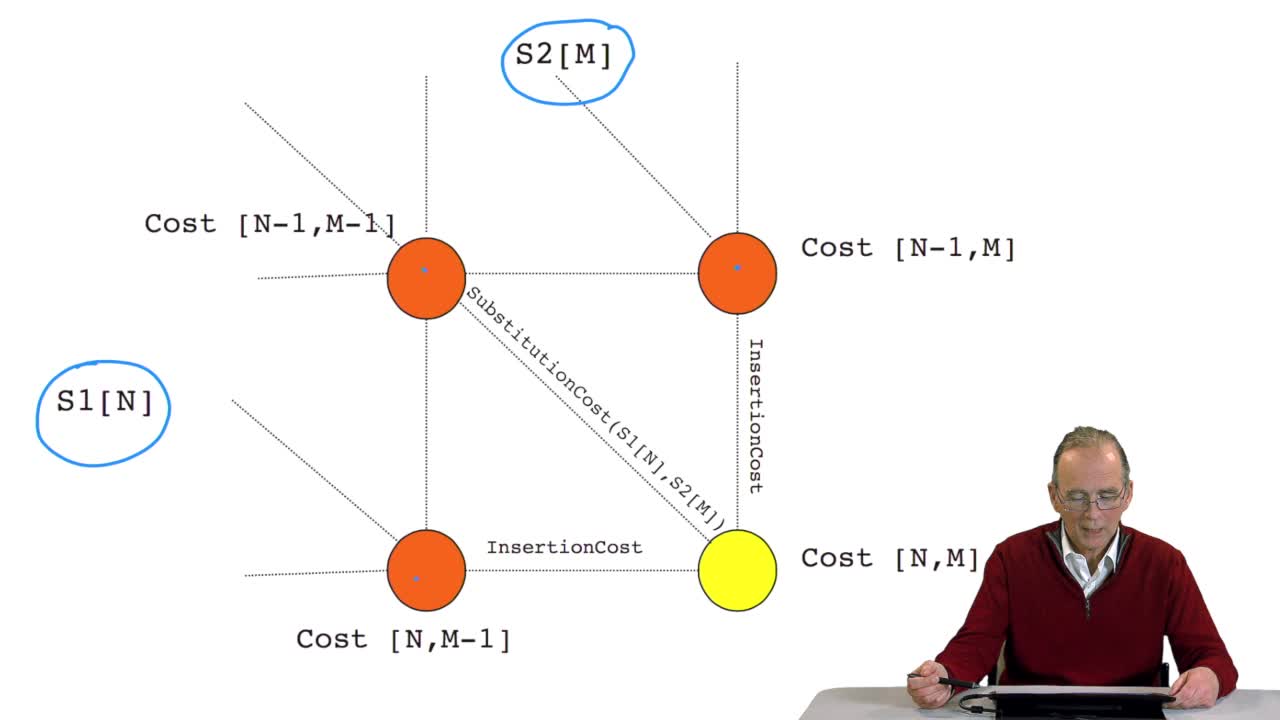
4.8. A recursive algorithm
We have seen how we can computethe optimal cost, the ending node of our grid if we know the optimal cost of the three adjacent nodes. This is this computation scheme we can see here using the notation
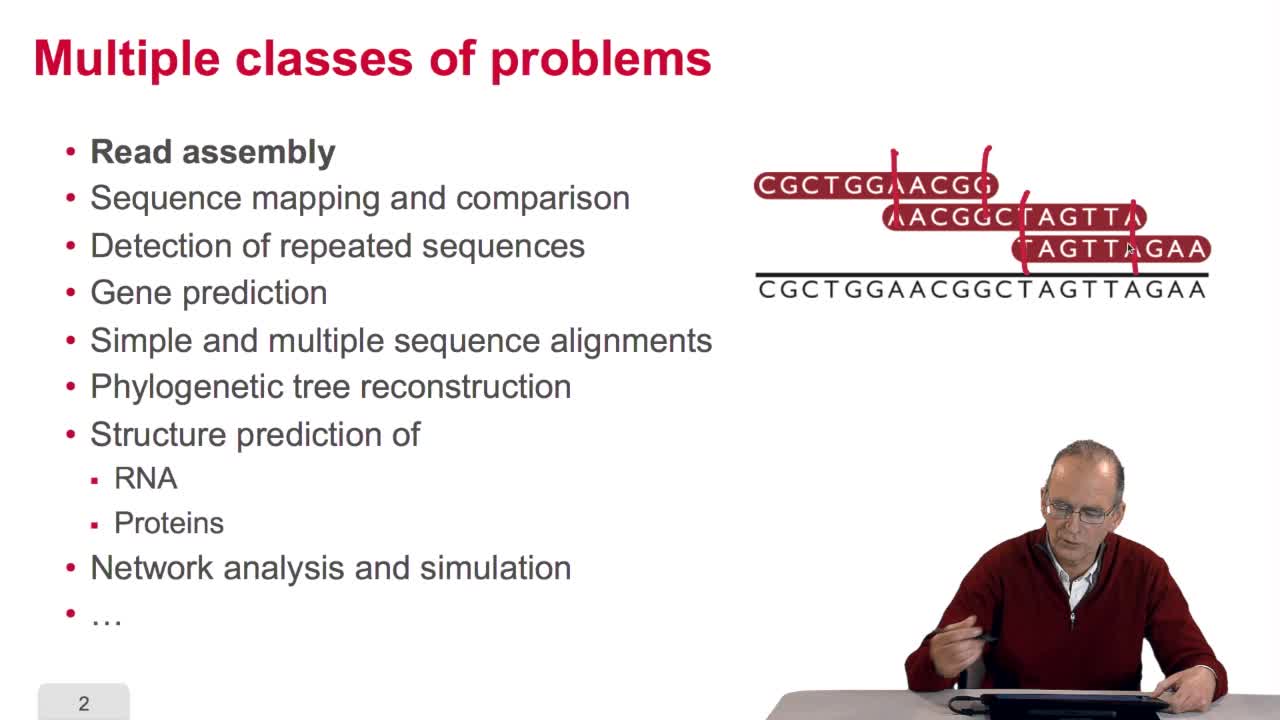
5.6. The diversity of bioinformatics algorithms
In this course, we have seen a very little set of bioinformatic algorithms. There exist numerous various algorithms in bioinformatics which deal with a large span of classes of problems. For example,