Rechenmann, François (19..-.... ; informaticien)
Ingénieur et Docteur-Ingénieur en informatique, François Rechenmann est chercheur au centre Inria Grenoble – Rhône-Alpes. Il y exerce ses activités à l’interface de l’informatique et des sciences du vivant en contribuant plus particulièrement au développement de méthodes et de logiciels pour l’analyse des séquences génomiques des microorganismes. Cofondateur de la société Genostar, qui propose des solutions bioinformatiques aux industries pharmaceutiques, agroalimentaires et biotechnologiques, il en est le conseiller scientifique. Très impliqué dans les actions de médiation scientifique, François Rechenmann est le responsable scientifique du site Interstices dont l’objectif est d’expliquer l’informatique en tant que domaine de recherche.
Vidéos
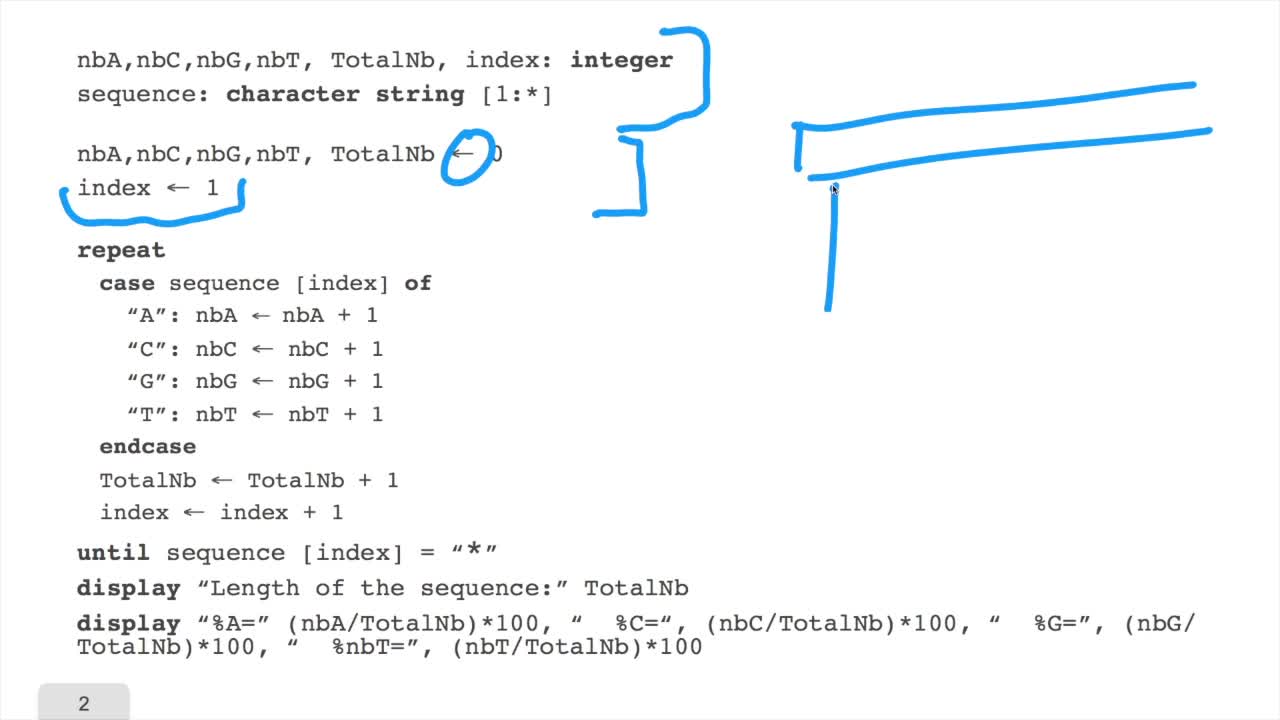
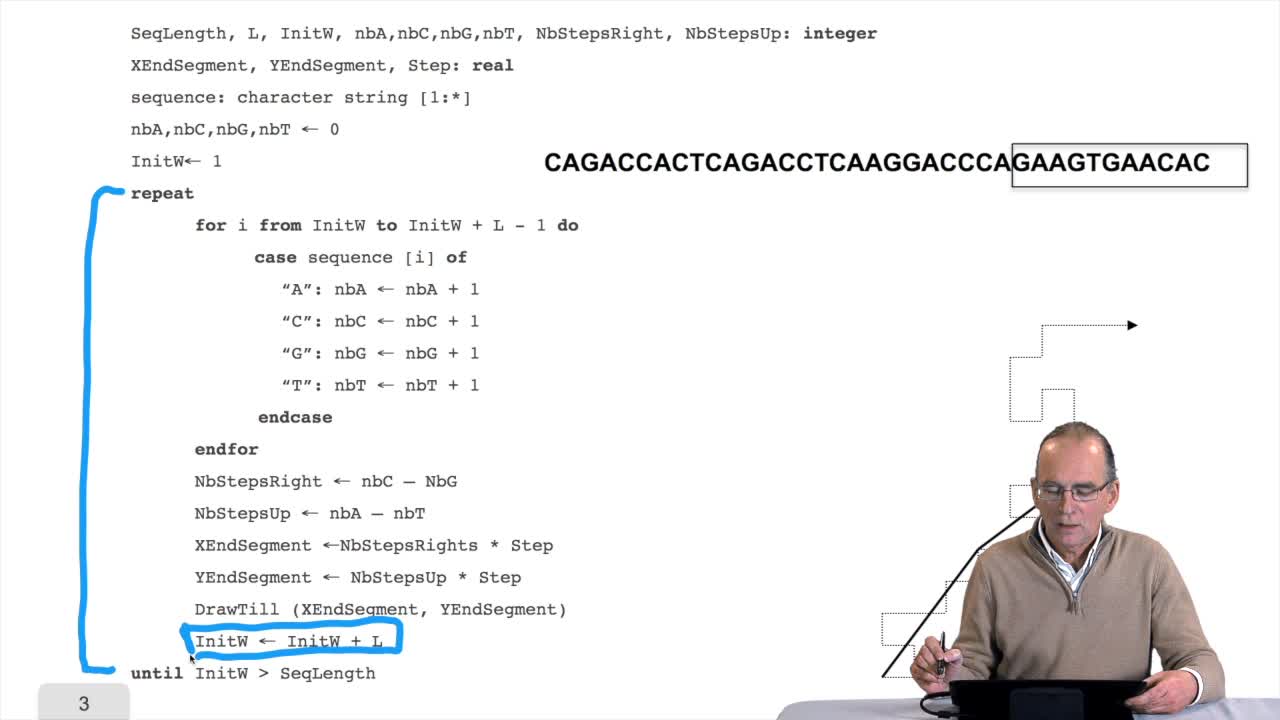
1.6. GC and AT contents of DNA sequence
We have designed our first algorithmfor counting nucleotides. Remember, what we have writtenin pseudo code is first declaration of variables. We have several integer variables that are variables which
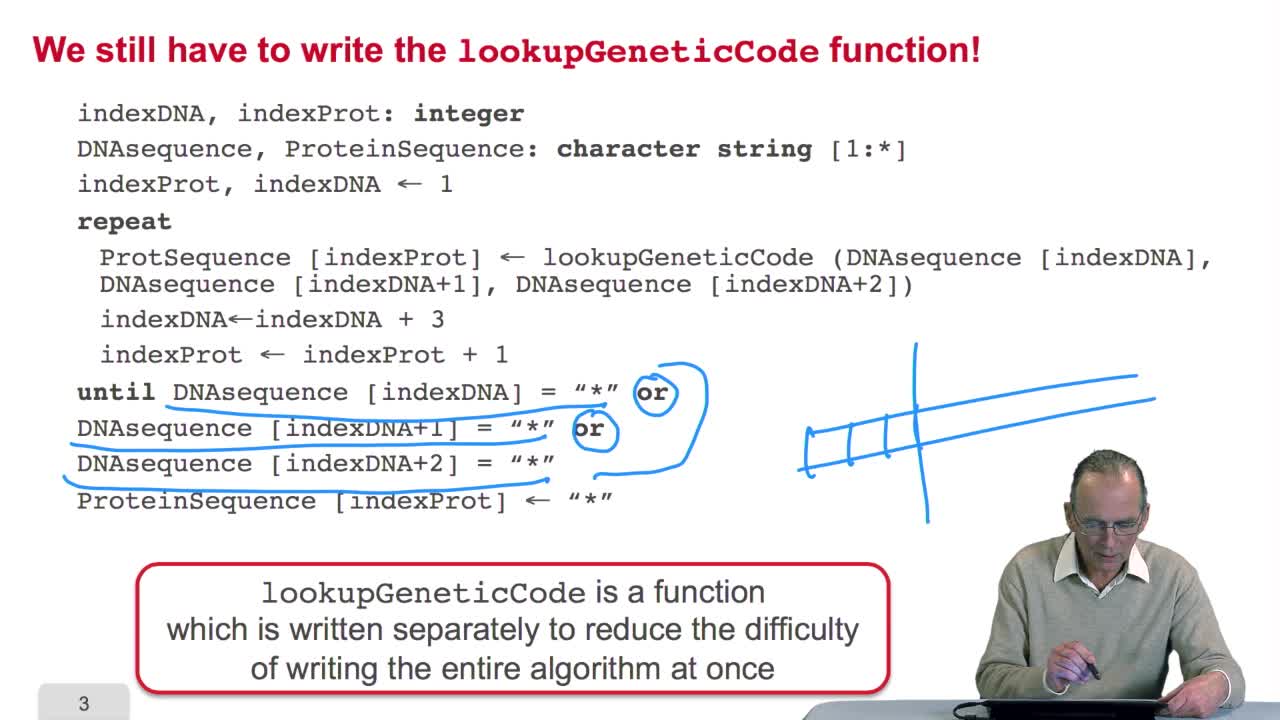
2.5. Implementing the genetic code
Remember we were designing our translation algorithm and since we are a bit lazy, we decided to make the hypothesis that there was the adequate function forimplementing the genetic code. It's now time
3.2. A simple algorithm for gene prediction
Based on the principle we statedin the last session, we will now write in pseudo code a firstalgorithm for locating genes on a bacterial genome. Remember first how this algorithm should work, we first
3.10. Gene prediction in eukaryotic genomes
If it is possible to have verygood predictions for bacterial genes, it's certainly not the caseyet for eukaryotic genomes. Eukaryotic cells have manydifferences in comparison to prokaryotic cells. You
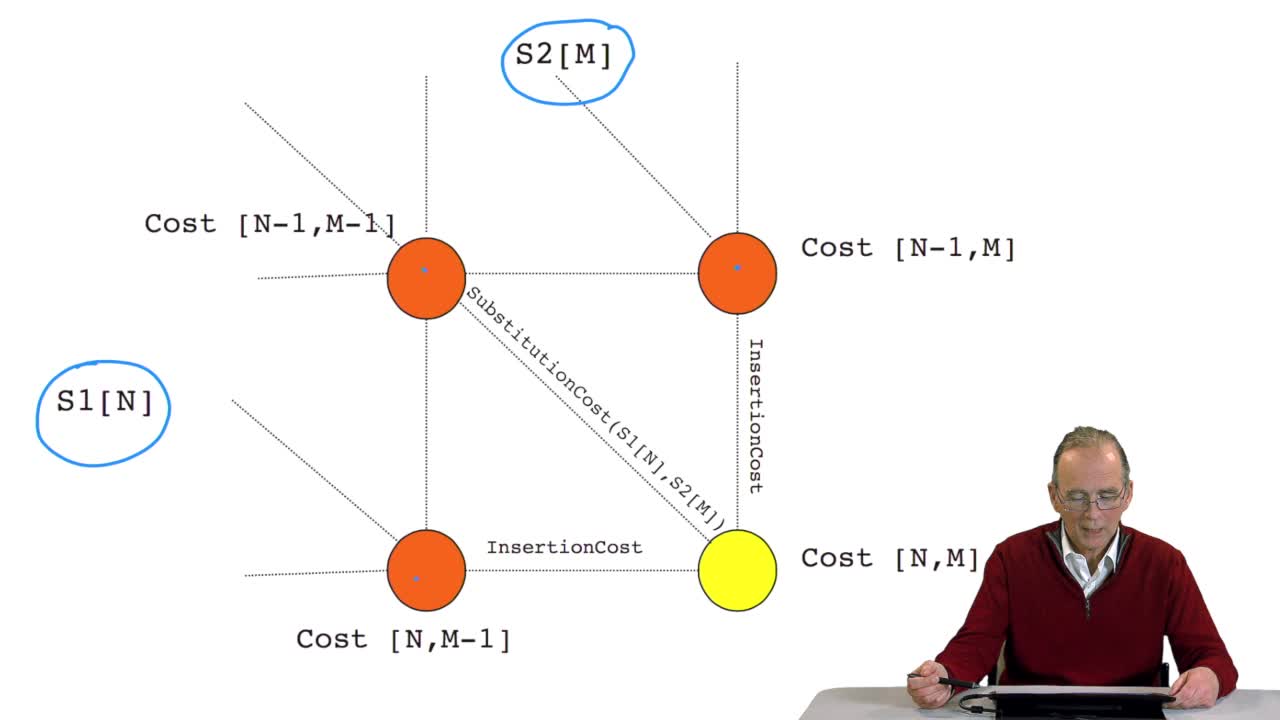
4.8. A recursive algorithm
We have seen how we can computethe optimal cost, the ending node of our grid if we know the optimal cost of the three adjacent nodes. This is this computation scheme we can see here using the notation
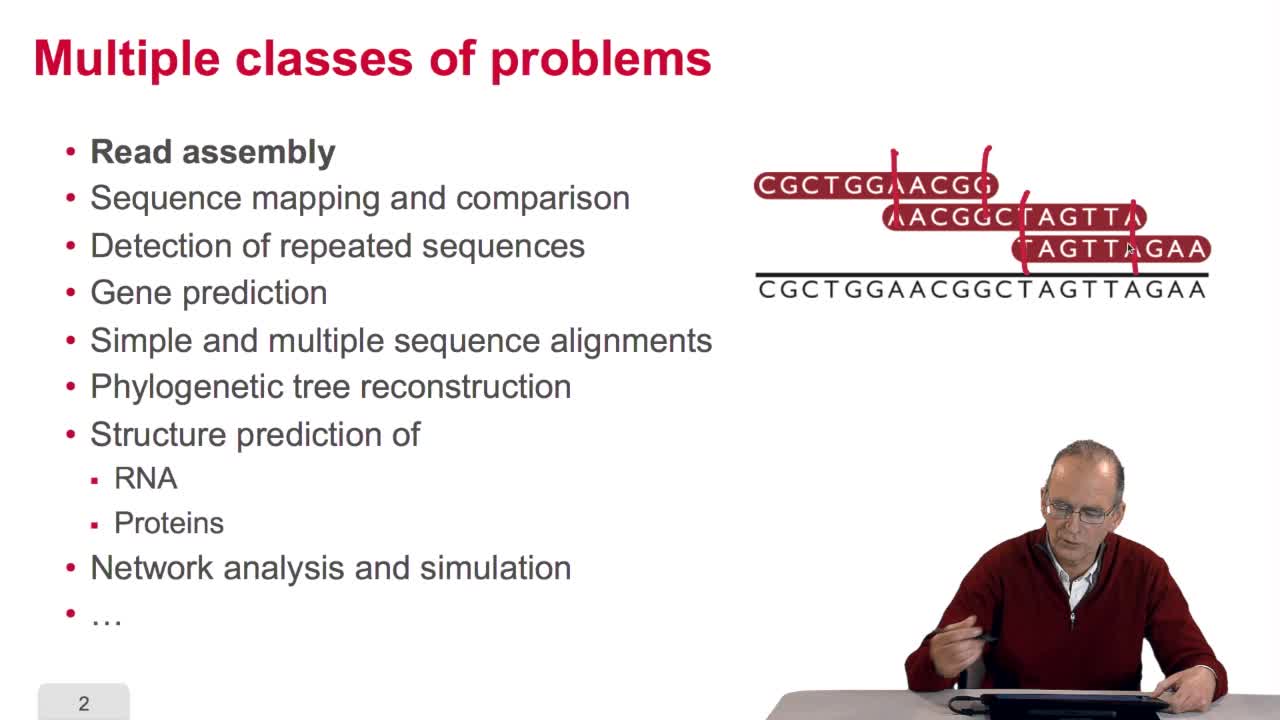
5.6. The diversity of bioinformatics algorithms
In this course, we have seen a very little set of bioinformatic algorithms. There exist numerous various algorithms in bioinformatics which deal with a large span of classes of problems. For example,
1.1. The cell, atom of the living world
Welcome to this introduction to bioinformatics. We will speak of genomes and algorithms. More specifically, we will see how genetic information can be analysed by algorithms. In these five weeks to
1.9. Predicting the origin of DNA replication?
We have seen a nice algorithm to draw, let's say, a DNA sequence. We will see that first, we have to correct a little bit this algorithm. And then we will see how such as imple algorithm can provide
2.8. DNA sequencing
During the last session, I explained several times how it was important to increase the efficiency of sequences processing algorithm because sequences arevery long and there are large volumes of
3.5. Making the predictions more reliable
We have got a bacterial gene predictor but the way this predictor works is rather crude and if we want to have more reliable results, we have to inject into this algorithmmore biological knowledge. We
4.6. A path is optimal if all its sub-paths are optimal
A sequence alignment between two sequences is a path in a grid. So that, an optimal sequence alignmentis an optimal path in the same grid. We'll see now that a property of this optimal path provides
5.1. The tree of life
Welcome to this fifth and last week of our course on genomes and algorithms that is the computer analysis of genetic information. During this week, we will firstsee what phylogenetic trees are and how