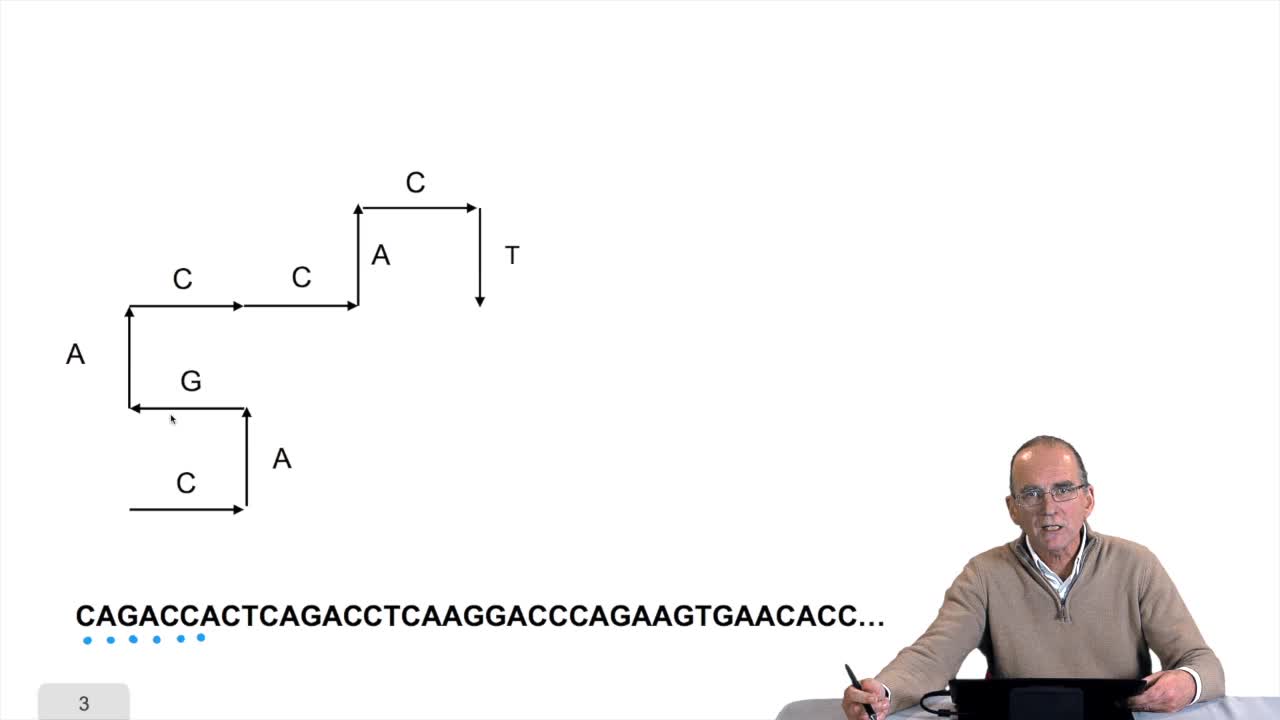1. Genomic texts

Descriptif
Table of contents
1.1. The cell, atom of the living world
1.2. At the heart of the cell: the DNA macromolecule
1.3. DNA codes for genetic information
1.4. What is an algorithm?
1.5. Counting nucleotides
1.6. GC and AT contents of DNA sequence
1.7. DNA walk
1.8. Compressing the DNA walk
1.9. Predicting the origin of DNA replication?
1.10. Overlapping sliding window
Vidéos
1.1. The cell, atom of the living world
Welcome to this introduction to bioinformatics. We will speak of genomes and algorithms. More specifically, we will see how genetic information can be analysed by algorithms. In these five weeks to
1.2. At the heart of the cell: the DNA macromolecule
During the last session, we saw how at the heart of the cell there's DNA in the nucleus, sometimes of cells, or directly in the cytoplasm of the bacteria. The DNA is what we call a macromolecule, that
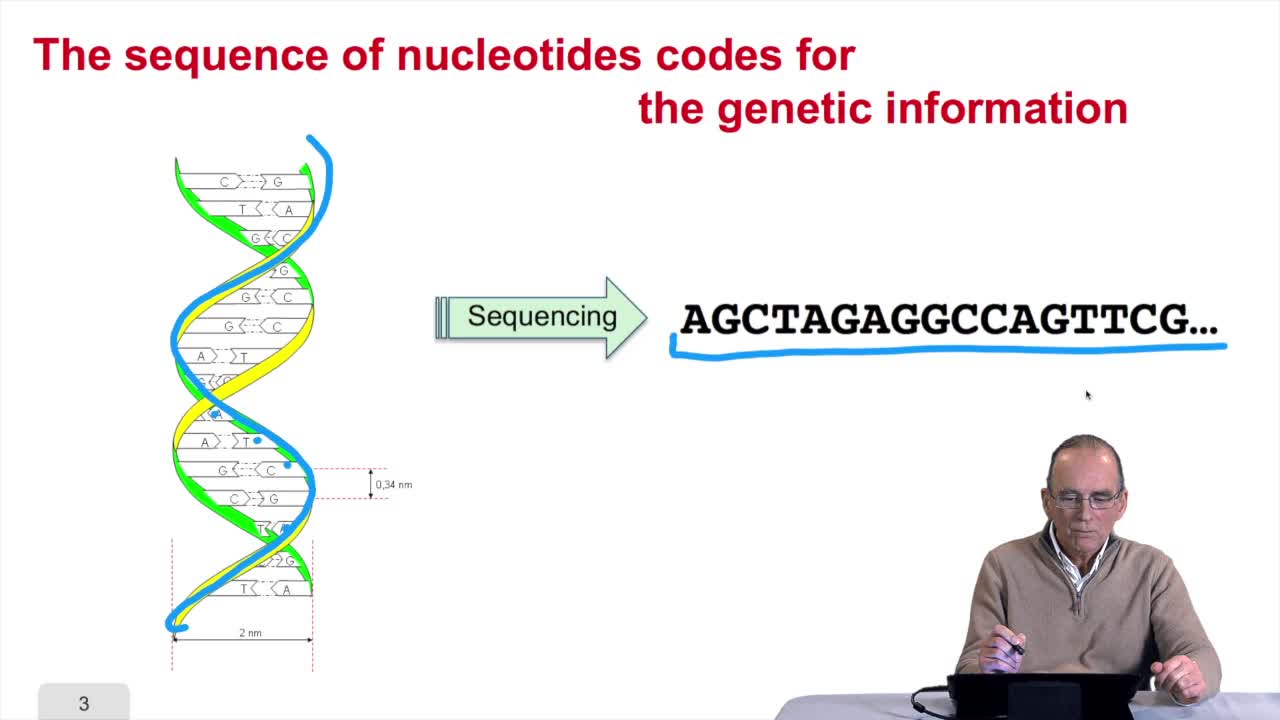
1.3. DNA codes for genetic information
Remember at the heart of any cell,there is this very long molecule which is called a macromolecule for this reason, which is the DNA molecule. Now we will see that DNA molecules support what is called
1.4. What is an algorithm?
We have seen that a genomic textcan be indeed a very long sequence of characters. And to interpret this sequence of characters, we will need to use computers. Using computers means writing program.
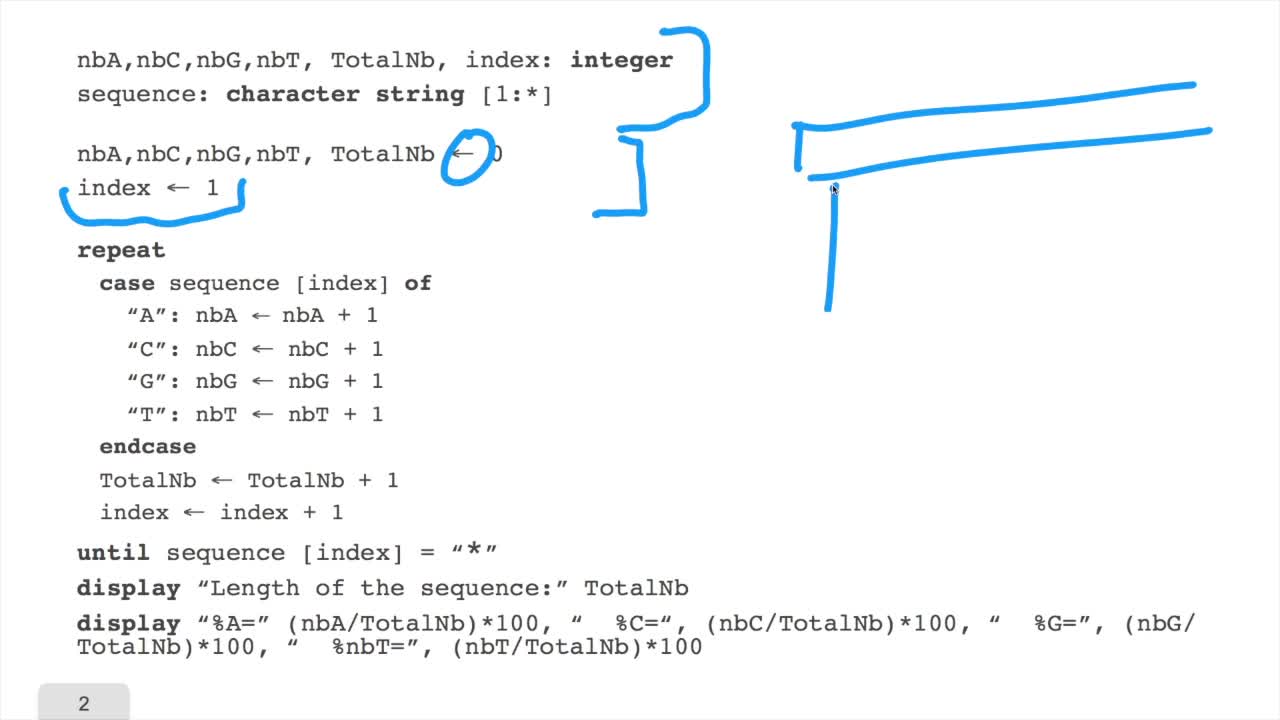
1.5. Counting nucleotides
In this session, don't panic. We will design our first algorithm. This algorithm is forcounting nucleotides. The idea here is that as an input,you have a sequence of nucleotides, of bases, of letters,
1.6. GC and AT contents of DNA sequence
We have designed our first algorithmfor counting nucleotides. Remember, what we have writtenin pseudo code is first declaration of variables. We have several integer variables that are variables which
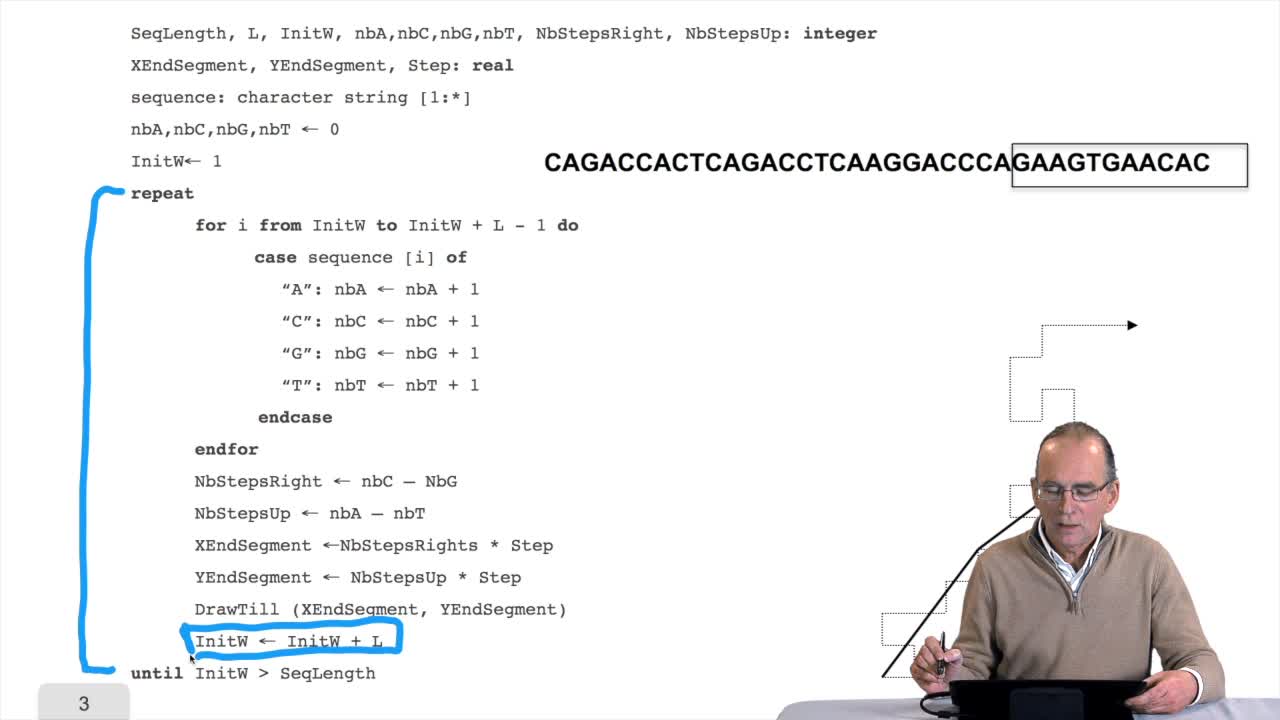
1.7. DNA walk
We will now design a more graphical algorithm which is called "the DNA walk". We shall see what does it mean "DNA walk". Walk on to DNA. Something like that, yes. But first, just have a look again at
1.8. Compressing the DNA walk
We have written the algorithm for the circle DNA walk. Just a precision here: the kind of drawing we get has nothing to do with the physical drawing of the DNA molecule. It is a symbolic
1.9. Predicting the origin of DNA replication?
We have seen a nice algorithm to draw, let's say, a DNA sequence. We will see that first, we have to correct a little bit this algorithm. And then we will see how such as imple algorithm can provide
1.10. Overlapping sliding window
We have made some drawings along a genomic sequence. And we have seen that although the algorithm is quite simple, even if some points of the algorithmare bit trickier than the others, we were able to
Intervenants et intervenantes
Ingénieur. Auteur d'une thèse de docteur-ingénieur en sciences appliquées (Grenoble INPG, 1976). - HDR. Directeur de thèse à Grenoble INPG (1990-1994-) et à l'université de Grenoble 1. Directeur de recherche au centre Inria Grenoble – Rhône-Alpes (2002, 2015)