3. Gene prediction

Descriptif
Table of contents
3.1. All genes end on a stop codon
3.2. A simple algorithm for gene prediction
3.3. Searching for start and stop codons
3.4. Predicting all the genes in a sequence
3.5. Making the predictions more reliable
3.6. Boyer-Moore algorithm
3.7. Index and suffix trees
3.8. Probabilistic methods
3.9. Benchmarking the prediction methods
3.10. Gene prediction in eukaryotic genomes
Vidéos
3.1. All genes end on a stop codon
Last week we studied genes and proteins and so how genes, portions of DNA, are translated into proteins. We also saw the very fast evolutionof the sequencing technology which allows for producing
3.2. A simple algorithm for gene prediction
Based on the principle we statedin the last session, we will now write in pseudo code a firstalgorithm for locating genes on a bacterial genome. Remember first how this algorithm should work, we first
3.3. Searching for start and stop codons
We have written an algorithm for finding genes. But you remember that we arestill to write the two functions for finding the next stop codonand the next start codon. Let's see how we can do that. We
3.4. Predicting all the genes in a sequence
We have written an algorithm whichis able to locate potential genes on a sequence but only on one phase because we are looking triplets after triplets. Now remember that the genes maybe located on
3.5. Making the predictions more reliable
We have got a bacterial gene predictor but the way this predictor works is rather crude and if we want to have more reliable results, we have to inject into this algorithmmore biological knowledge. We
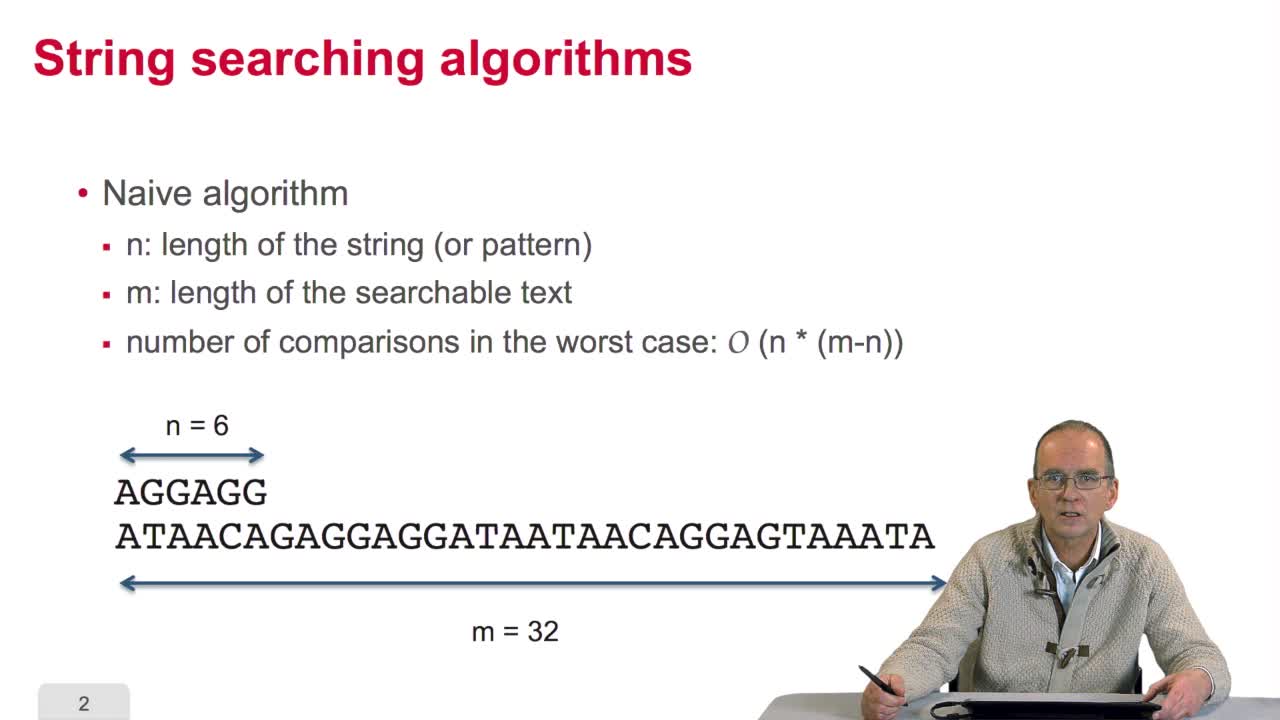
3.6. Boyer-Moore algorithm
We have seen how we can make gene predictions more reliable through searching for all the patterns,all the occurrences of patterns. We have seen, for example, howif we locate the RBS, Ribosome
3.7. Index and suffix trees
We have seen with the Boyer-Moore algorithm how we can increase the efficiency of spin searching through the pre-processing of the pattern to be searched. Now we will see that an alternative way of
3.8. Probabilistic methods
Up to now, to predict our gene,we only rely on the process of searching certain strings or patterns. In order to further improve our gene predictor, the idea is to use, to rely onprobabilistic methods
3.9. Benchmarking the prediction methods
It is necessary to underline that gene predictors produce predictions. Predictions mean that you have no guarantees that the coding sequences, the coding regions,the genes you get when applying your
3.10. Gene prediction in eukaryotic genomes
If it is possible to have verygood predictions for bacterial genes, it's certainly not the caseyet for eukaryotic genomes. Eukaryotic cells have manydifferences in comparison to prokaryotic cells. You
Intervenants et intervenantes
Ingénieur. Auteur d'une thèse de docteur-ingénieur en sciences appliquées (Grenoble INPG, 1976). - HDR. Directeur de thèse à Grenoble INPG (1990-1994-) et à l'université de Grenoble 1. Directeur de recherche au centre Inria Grenoble – Rhône-Alpes (2002, 2015)