5. Phylogenetic trees
Descriptif
Table of contents
5.1. The tree of life
5.2. The tree, an abstract object
5.3. Building an array of distances
5.4. The UPGMA algorithm
5.5. Differences are not always what they look like
5.6. The diversity of bioinformatics algorithms
5.7. The application domains in microbiology
Vidéos
5.1. The tree of life
Welcome to this fifth and last week of our course on genomes and algorithms that is the computer analysis of genetic information. During this week, we will firstsee what phylogenetic trees are and how
5.2. The tree, an abstract object
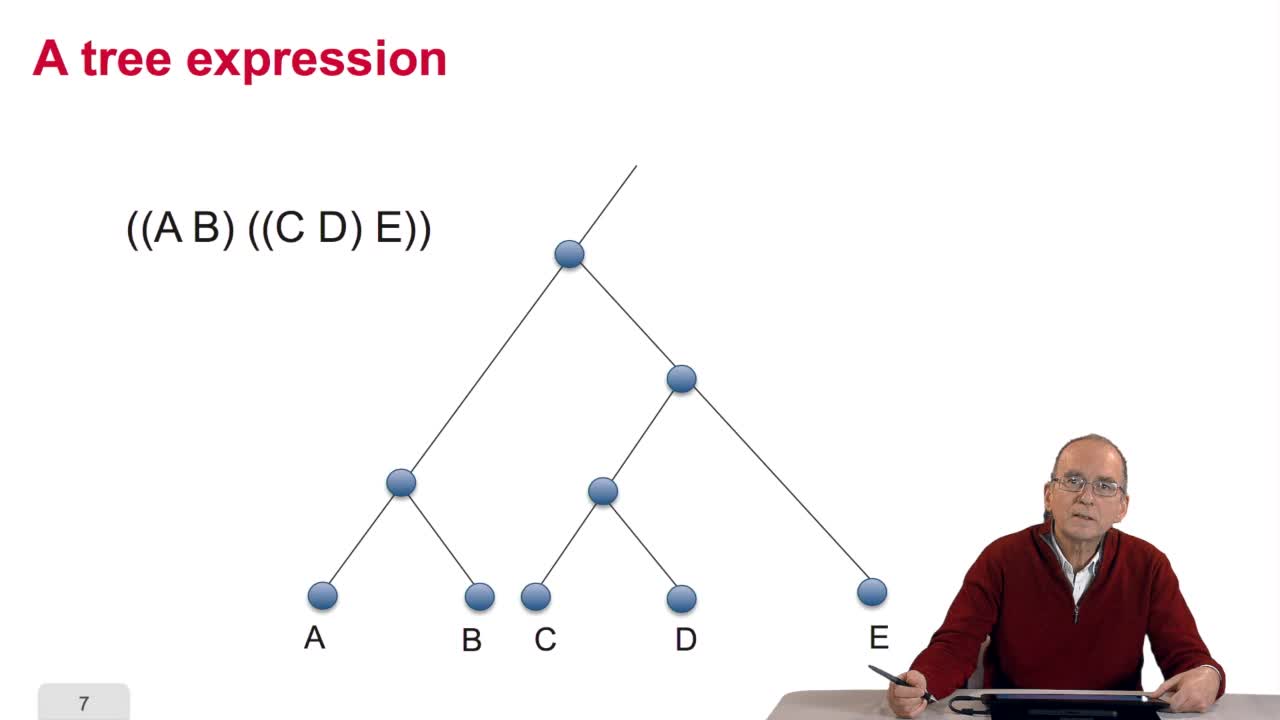
When we speak of trees, of species,of phylogenetic trees, of course, it's a metaphoric view of a real tree. Our trees are abstract objects. Here is a tree and the different components of this tree.
5.3. Building an array of distances
So using the sequences of homologous gene between several species, our aim is to reconstruct phylogenetic tree of the corresponding species. For this, we have to comparesequences and compute distances
5.4. The UPGMA algorithm
We know how to fill an array with the values of the distances between sequences, pairs of sequences which are available in the file. This array of distances will be the input of our algorithm for
5.5. Differences are not always what they look like
The algorithm we have presented works on an array of distance between sequences. These distances are evaluated on the basis of differences between the sequences. The problem is that behind the
5.6. The diversity of bioinformatics algorithms
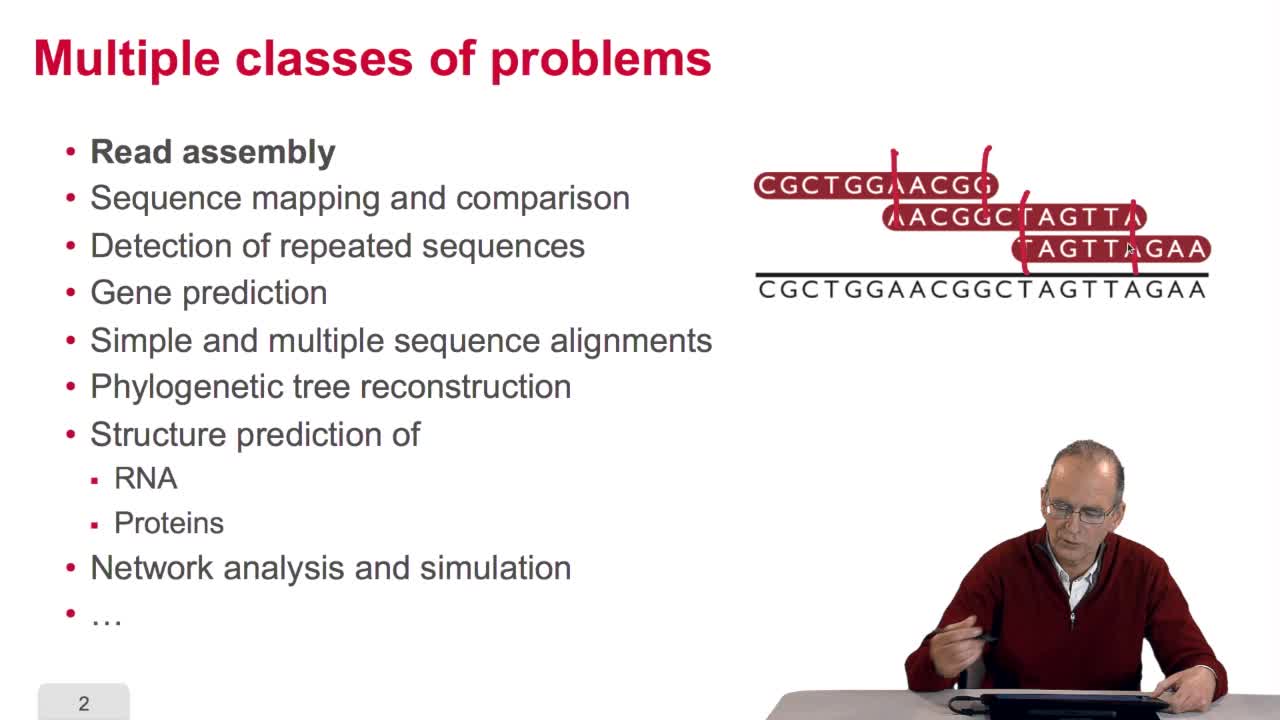
In this course, we have seen a very little set of bioinformatic algorithms. There exist numerous various algorithms in bioinformatics which deal with a large span of classes of problems. For example,
5.7. The application domains in microbiology
Bioinformatics relies on many domains of mathematics and computer science. Of course, algorithms themselves on character strings are important in bioinformatics, we have seen them. Algorithms and
Intervenants et intervenantes
Ingénieur. Auteur d'une thèse de docteur-ingénieur en sciences appliquées (Grenoble INPG, 1976). - HDR. Directeur de thèse à Grenoble INPG (1990-1994-) et à l'université de Grenoble 1. Directeur de recherche au centre Inria Grenoble – Rhône-Alpes (2002, 2015)